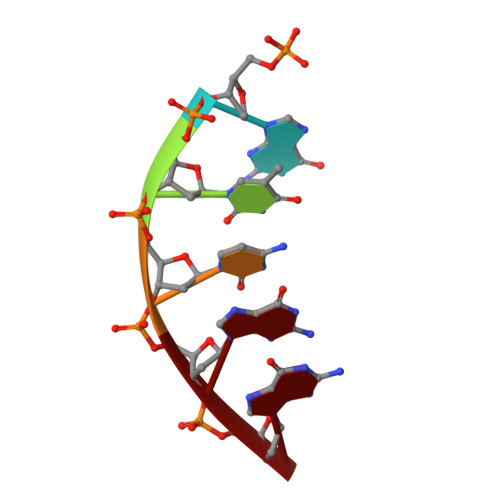
Transition-state destabilization reveals how human DNA polymerase beta proceeds across the chemically unstable lesion N7-methylguanine.
Koag, M.C., Kou, Y., Ouzon-Shubeita, H., Lee, S.(2014) Nucleic Acids Res 42: 8755-8766
- PubMed: 24966350
- DOI: https://doi.org/10.1093/nar/gku554
- Primary Citation of Related Structures:
4O5C, 4O5E, 4O5K, 4P2H - PubMed Abstract:
N7-Methyl-2'-deoxyguanosine (m7dG) is the predominant lesion formed by methylating agents. A systematic investigation on the effect of m7dG on DNA replication has been difficult due to the chemical instability of m7dG. To gain insights into the m7dG effect, we employed a 2'-fluorine-mediated transition-state destabilzation strategy. Specifically, we determined kinetic parameters for dCTP insertion opposite a chemically stable m7dG analogue, 2'-fluoro-m7dG (Fm7dG), by human DNA polymerase β (polβ) and solved three X-ray structures of polβ in complex with the templating Fm7dG paired with incoming dCTP or dTTP analogues. The kinetic studies reveal that the templating Fm7dG slows polβ catalysis ∼ 300-fold, suggesting that m7dG in genomic DNA may impede replication by some DNA polymerases. The structural analysis reveals that Fm7dG forms a canonical Watson-Crick base pair with dCTP, but metal ion coordination is suboptimal for catalysis in the polβ-Fm7dG:dCTP complex, which partially explains the slow insertion of dCTP opposite Fm7dG by polβ. In addition, the polβ-Fm7dG:dTTP structure shows open protein conformations and staggered base pair conformations, indicating that N7-methylation of dG does not promote a promutagenic replication. Overall, the first systematic studies on the effect of m7dG on DNA replication reveal that polβ catalysis across m7dG is slow, yet highly accurate.
Organizational Affiliation:
Division of Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin, Austin, TX 78712, USA SeongminLee@austin.utexas.edu.