Ubiquitin-specific protease 7 is a regulator of ubiquitin-conjugating enzyme UbE2E1.
Sarkari, F., Wheaton, K., La Delfa, A., Mohamed, M., Shaikh, F., Khatun, R., Arrowsmith, C.H., Frappier, L., Saridakis, V., Sheng, Y.(2013) J Biol Chem 288: 16975-16985
- PubMed: 23603909
- DOI: https://doi.org/10.1074/jbc.M113.469262
- Primary Citation of Related Structures:
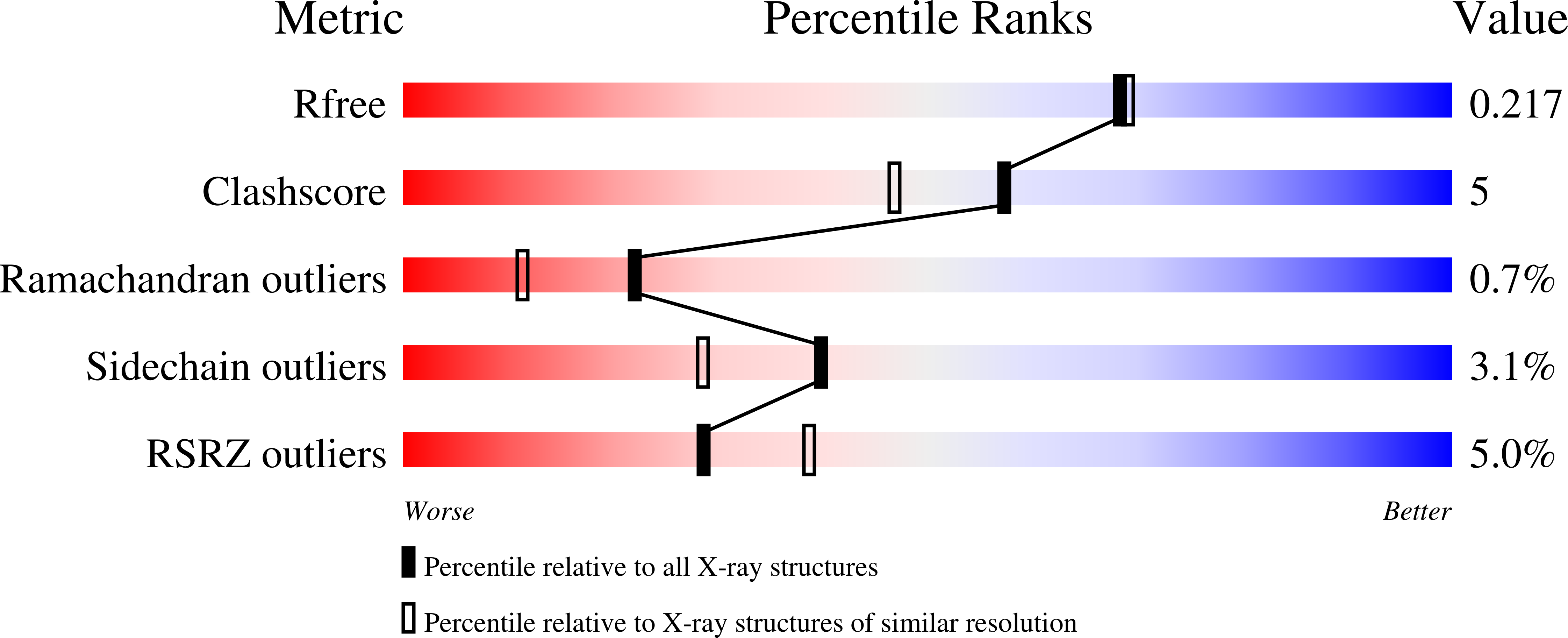
4JJQ - PubMed Abstract:
Ubiquitin-specific protease 7 (USP7) is a deubiquitinating enzyme found in all eukaryotes that catalyzes the removal of ubiquitin from specific target proteins. Here, we report that UbE2E1, an E2 ubiquitin conjugation enzyme with a unique N-terminal extension, is a novel USP7-interacting protein. USP7 forms a complex with UbE2E1 in vitro and in vivo through the ASTS USP7 binding motif within its N-terminal extension in an identical manner with other known USP7 binding proteins. We show that USP7 attenuates UbE2E1-mediated ubiquitination, an effect that requires the N-terminal ASTS sequence of UbE2E1 as well as the catalytic activity of USP7. Additionally, USP7 is critical in maintaining the steady state levels of UbE2E1 in cells. This study reveals a new cellular mechanism that couples the opposing activities of the ubiquitination machinery and a deubiquitinating enzyme to maintain and modulate the dynamic balance of the ubiquitin-proteasome system.
Organizational Affiliation:
Department of Biology, York University, Toronto, Ontario M3J 1P3.