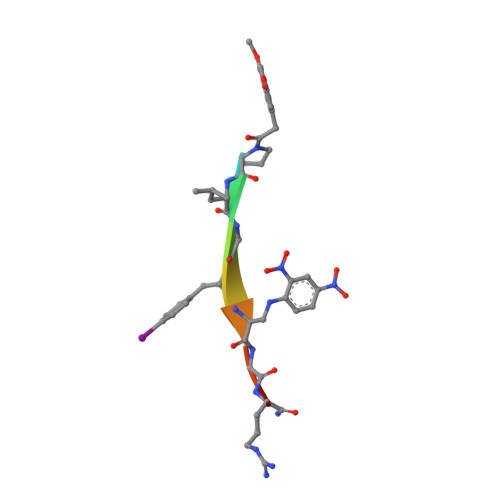
Halogen Bonding Controls Selectivity of FRET Substrate Probes for MMP-9.
Tranchant, I., Vera, L., Czarny, B., Amoura, M., Cassar, E., Beau, F., Stura, E.A., Dive, V.(2014) Chem Biol 21: 408-413
- PubMed: 24583051
- DOI: https://doi.org/10.1016/j.chembiol.2014.01.008
- Primary Citation of Related Structures:
4JIJ, 4JQG - PubMed Abstract:
Matrix metalloproteinases (MMPs) are a large family of zinc-dependent endoproteases that catalyze cleavage of extracellular matrix and nonmatrix proteins. MMPs play a role in tissue remodeling, and their uncontrolled activity is associated with number of diseases, including tumor metastasis. Thus, there is a need to develop methods to monitor MMP activity, and number of probes has been previously described. The key problem many probes encounter is the issue of selectivity, since 23 human MMPs, despite playing different physiological roles, have structurally similar active sites. Here, we introduce the halogen bonding concept into the probe design and show that the probe containing iodine exhibits an unprecedented selectivity for MMP-9. We provide structure-based explanation for the selectivity, confirming that it is due to formation of the halogen bond that supports catalysis, and we highlight the value of exploring halogen bonding in the context of selective probe design.
Organizational Affiliation:
CEA, iBiTec-S, Service d'Ingénierie Moléculaire des Protéines (SIMOPRO), Labex LERMIT, CE-Saclay, 91191 Gif sur Yvette Cedex, France.