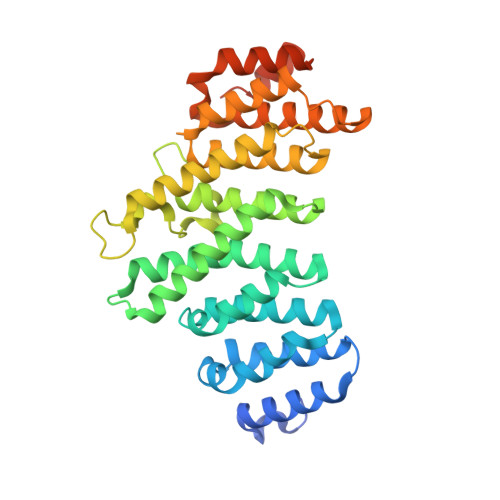
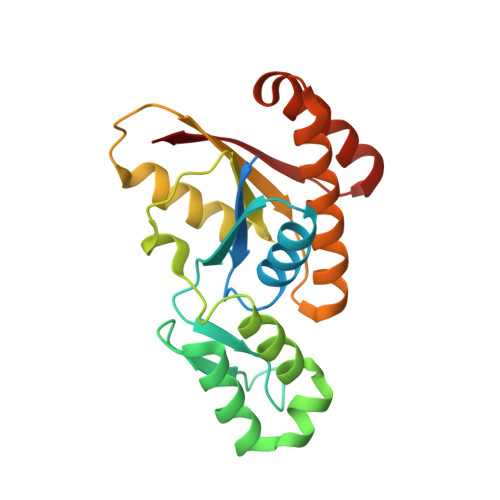
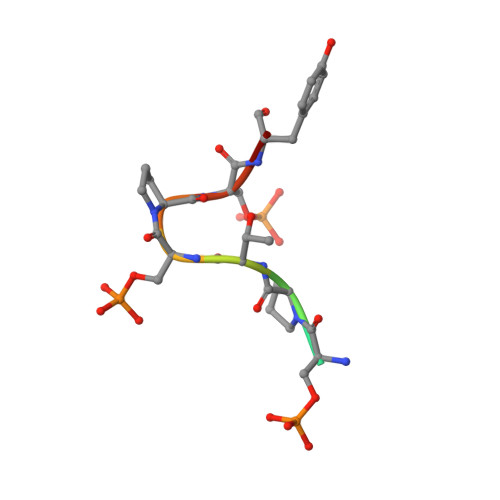
An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72.
Xiang, K., Manley, J.L., Tong, L.(2012) Genes Dev 26: 2265-2270
- PubMed: 23070812
- DOI: https://doi.org/10.1101/gad.198853.112
- Primary Citation of Related Structures:
4H3H, 4H3K - PubMed Abstract:
Ssu72, an RNA polymerase II C-terminal domain (CTD) phospho-Ser5 (pSer5) phosphatase, was recently reported to have pSer7 phosphatase activity as well. We report here the crystal structure of a ternary complex of the N-terminal domain of human symplekin, human Ssu72, and a 10-mer pSer7 CTD peptide. Surprisingly, the peptide is bound in the Ssu72 active site with its backbone running in the opposite direction compared with a pSer5 peptide. The pSer7 phosphatase activity of Ssu72 is ∼4000-fold lower than its pSer5 phosphatase activity toward a peptide substrate, consistent with the structural observations.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York 10027, USA.