High resolution crystal structures of Carbonic Anhydrase II in complex with novel sulfamide binders
Di Pizio, A., Schulze Wischeler, J., Haake, M., Heine, A., Supuran, C., Klebe, G.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Carbonic anhydrase 2 | 260 | Homo sapiens | Mutation(s): 0 Gene Names: CA2 EC: 4.2.1.1 (PDB Primary Data), 4.2.1.69 (UniProt) | 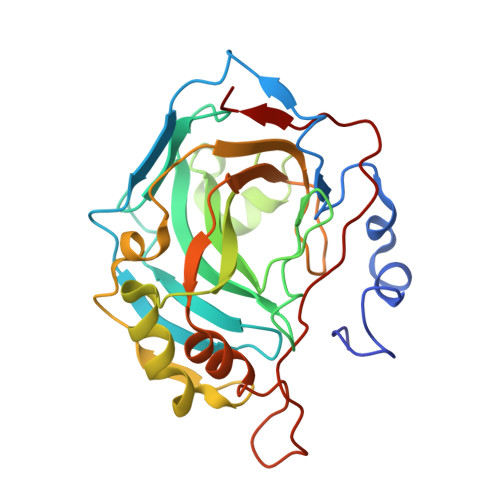 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P00918 (Homo sapiens) Explore P00918 Go to UniProtKB: P00918 | |||||
PHAROS: P00918 GTEx: ENSG00000104267 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00918 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MBO Query on MBO | B [auth A] | MERCURIBENZOIC ACID C7 H5 Hg O2 FVFZSVRSDNUCGG-UHFFFAOYSA-N |  | ||
| 0VV Query on 0VV | D [auth A], E [auth A] | N-[(2E)-3,4-dihydroquinazolin-2(1H)-ylidene]sulfuric diamide C8 H10 N4 O2 S NQQBFHZTZMKZCO-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | F [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| DMS Query on DMS | G [auth A] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | C [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.139 | α = 90 |
| b = 41.457 | β = 104.21 |
| c = 72.169 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| PHASER | phasing |
| SHELXL-97 | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |