Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
Thompson, J.W., Salahudeen, A.A., Chollangi, S., Ruiz, J.C., Brautigam, C.A., Makris, T.M., Lipscomb, J.D., Tomchick, D.R., Bruick, R.K.(2012) J Biol Chem 287: 7357-7365
- PubMed: 22253436
- DOI: https://doi.org/10.1074/jbc.M111.308684
- Primary Citation of Related Structures:
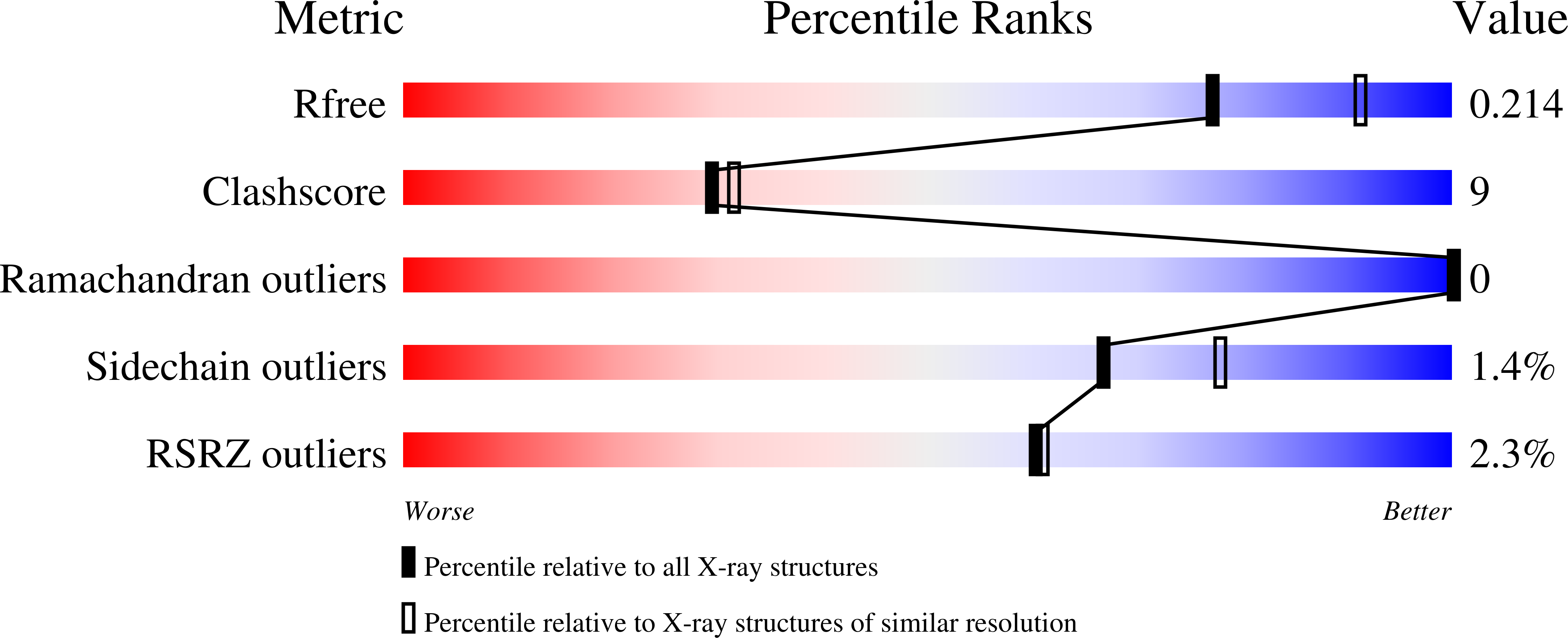
3V5X, 3V5Y, 3V5Z - PubMed Abstract:
Mammalian cells maintain iron homeostasis by sensing changes in bioavailable iron levels and promoting adaptive responses. FBXL5 is a subunit of an E3 ubiquitin ligase complex that mediates the stability of iron regulatory protein 2, an important posttranscriptional regulator of several genes involved in iron metabolism. The stability of FBXL5 is regulated in an iron- and oxygen-responsive manner, contingent upon the presence of its N-terminal domain. Here we present the atomic structure of the FBXL5 N terminus, a hemerythrin-like α-helical bundle fold not previously observed in mammalian proteins. The core of this domain employs an unusual assortment of amino acids necessary for the assembly and sensing properties of its diiron center. These regulatory features govern the accessibility of a mapped sequence required for proteasomal degradation of FBXL5. Detailed molecular and structural characterization of the ligand-responsive hemerythrin domain provides insights into the mechanisms by which FBXL5 serves as a unique mammalian metabolic sensor.
Organizational Affiliation:
Department of Biochemistry, University of Texas Southwestern Medical Center at Dallas, Dallas, Texas 75390, USA.