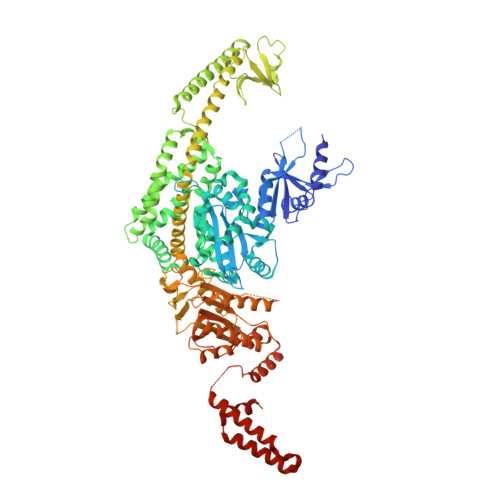
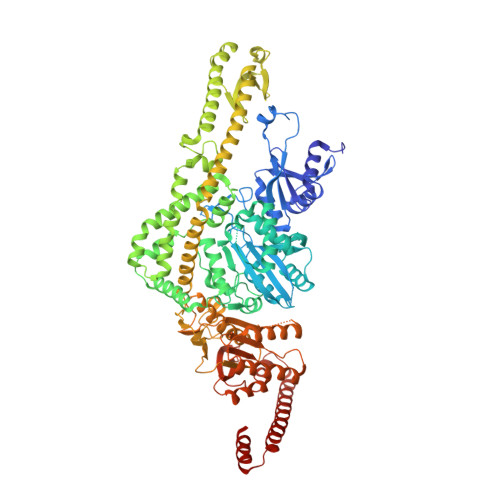
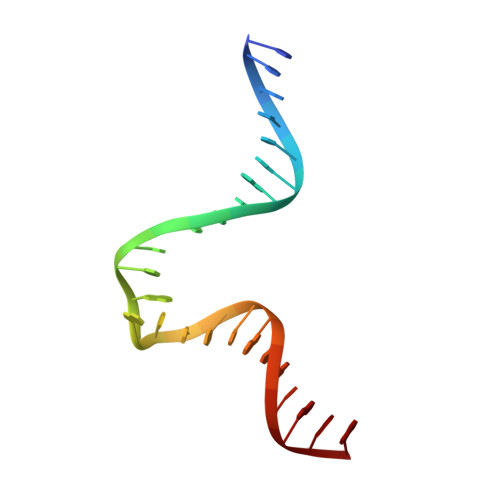
Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops
Gupta, S., Gellert, M., Yang, W.(2012) Nat Struct Mol Biol 19: 72-78
- PubMed: 22179786
- DOI: https://doi.org/10.1038/nsmb.2175
- Primary Citation of Related Structures:
3THW, 3THX, 3THY, 3THZ - PubMed Abstract:
DNA mismatch repair corrects replication errors, thus reducing mutation rates and microsatellite instability. Genetic defects in this pathway cause Lynch syndrome and various cancers in humans. Binding of a mispaired or unpaired base by bacterial MutS and eukaryotic MutSα is well characterized. We report here crystal structures of human MutSβ in complex with DNA containing insertion-deletion loops (IDL) of two, three, four or six unpaired nucleotides. In contrast to eukaryotic MutSα and bacterial MutS, which bind the base of a mismatched nucleotide, MutSβ binds three phosphates in an IDL. DNA is severely bent at the IDL; unpaired bases are flipped out into the major groove and partially exposed to solvent. A normal downstream base pair can become unpaired; a single unpaired base can thereby be converted to an IDL of two nucleotides and recognized by MutSβ. The C-terminal dimerization domains form an integral part of the MutS structure and coordinate asymmetrical ATP hydrolysis by Msh2 and Msh3 with mismatch binding to signal for repair.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, US National Institutes of Health, Bethesda, Maryland, USA.