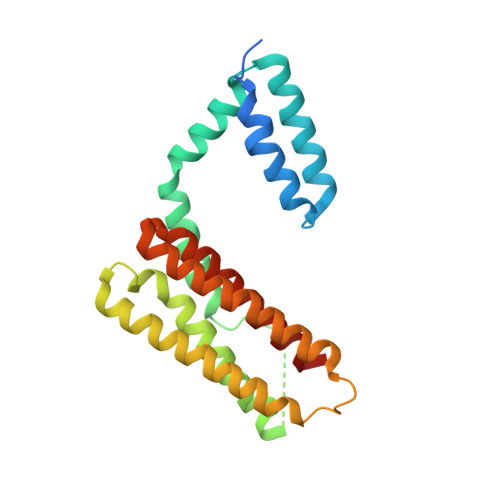
Molecular Recognition of Leucine-Aspartate Repeat (LD) Motifs by the Focal Adhesion Targeting Homology Domain of Cerebral Cavernous Malformation 3 (CCM3).
Li, X., Ji, W., Zhang, R., Folta-Stogniew, E., Min, W., Boggon, T.J.(2011) J Biol Chem 286: 26138-26147
- PubMed: 21632544
- DOI: https://doi.org/10.1074/jbc.M110.211250
- Primary Citation of Related Structures:
3RQE, 3RQF, 3RQG - PubMed Abstract:
Cerebral cavernous malformation (CCM) is a disease that affects between 0.1 and 0.5% of the human population, with mutations in CCM3 accounting for ~ 15% of the autosomal dominant form of the disease. We recently reported that CCM3 contains an N-terminal dimerization domain (CCM3D) and a C-terminal focal adhesion targeting (FAT) homology domain. Intermolecular protein-protein interactions of CCM3 are mediated by a highly conserved surface on the FAT homology domain and are affected by CCM3 truncations in the human disease. Here we report the crystal structures of CCM3 in complex with three different leucine-aspartate repeat (LD) motifs (LD1, LD2, and LD4) from the scaffolding protein paxillin, at 2.8, 2.7, and 2.5 Å resolution. We show that CCM3 binds LD motifs using the highly conserved hydrophobic patch 1 (HP1) and that this binding is similar to the binding of focal adhesion kinase and Pyk2 FAT domains to paxillin LD motifs. We further show by surface plasmon resonance that CCM3 binds paxillin LD motifs with affinities in the micromolar range, similar to FAK family FAT domains. Finally, we show that endogenous CCM3 and paxillin co-localize in mouse cerebral pericytes. These studies provide a molecular-level framework to investigate the protein-protein interactions of CCM3.
Organizational Affiliation:
Department of Pharmacology, Yale University School ofMedicine, New Haven, Connecticut 06520, USA.