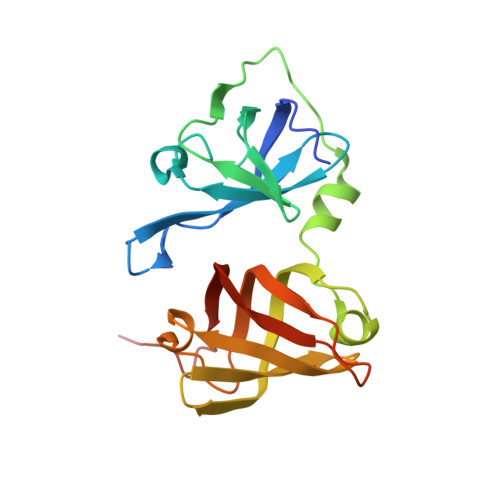
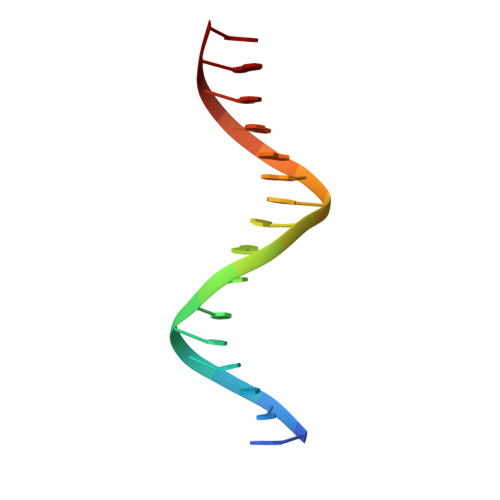
Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Jin, T., Perry, A., Jiang, J., Smith, P., Curry, J.A., Unterholzner, L., Jiang, Z., Horvath, G., Rathinam, V.A., Johnstone, R.W., Hornung, V., Latz, E., Bowie, A.G., Fitzgerald, K.A., Xiao, T.S.(2012) Immunity 36: 561-571
- PubMed: 22483801
- DOI: https://doi.org/10.1016/j.immuni.2012.02.014
- Primary Citation of Related Structures:
3RLN, 3RLO, 3RN2, 3RN5, 3RNU - PubMed Abstract:
Recognition of DNA by the innate immune system is central to antiviral and antibacterial defenses, as well as an important contributor to autoimmune diseases involving self DNA. AIM2 (absent in melanoma 2) and IFI16 (interferon-inducible protein 16) have been identified as DNA receptors that induce inflammasome formation and interferon production, respectively. Here we present the crystal structures of their HIN domains in complex with double-stranded (ds) DNA. Non-sequence-specific DNA recognition is accomplished through electrostatic attraction between the positively charged HIN domain residues and the dsDNA sugar-phosphate backbone. An intramolecular complex of the AIM2 Pyrin and HIN domains in an autoinhibited state is liberated by DNA binding, which may facilitate the assembly of inflammasomes along the DNA staircase. These findings provide mechanistic insights into dsDNA as the activation trigger and oligomerization platform for the assembly of large innate signaling complexes such as the inflammasomes.
Organizational Affiliation:
Structural Immunobiology Unit, Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892-0430, USA.