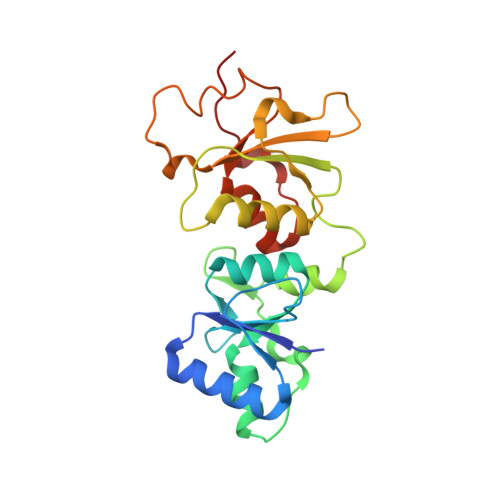
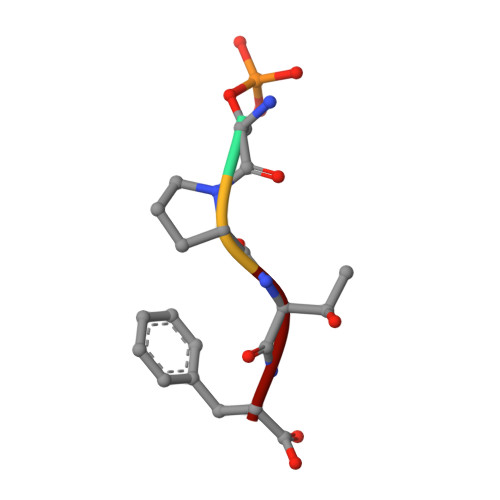
Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Campbell, S.J., Edwards, R.A., Glover, J.N.(2010) Structure 18: 167-176
- PubMed: 20159462
- DOI: https://doi.org/10.1016/j.str.2009.12.008
- Primary Citation of Related Structures:
3K05, 3K0H, 3K0K, 3K15, 3K16 - PubMed Abstract:
The tandem BRCT domains of BRCA1 and MDC1 facilitate protein signaling at DNA damage foci through specific interactions with serine-phosphorylated protein partners. The MDC1 BRCT binds pSer-Gln-Glu-Tyr-COO(-) at the C terminus of the histone variant gammaH2AX via direct recognition of the C-terminal carboxylate, while BRCA1 recognizes pSer-X-X-Phe motifs either at C-terminal or internal sites within target proteins. Using fluorescence polarization binding assays, we show that while both BRCTs prefer a free main chain carboxylate at the +3 position, this preference is much more pronounced in MDC1. Crystal structures of BRCA1 and MDC1 bound to tetrapeptide substrates reveal differences in the environment of conserved arginines (Arg1699 in BRCA1 and Arg1933 in MDC1) that determine the relative affinity for peptides with -COO(-) versus -CO-NH(2) termini. A mutation in MDC1 that induces a more BRCA1-like conformation relaxes the binding specificity, allowing the mutant to bind phosphopeptides lacking a -COO(-) terminus.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, AB T6G 2H7, Canada.