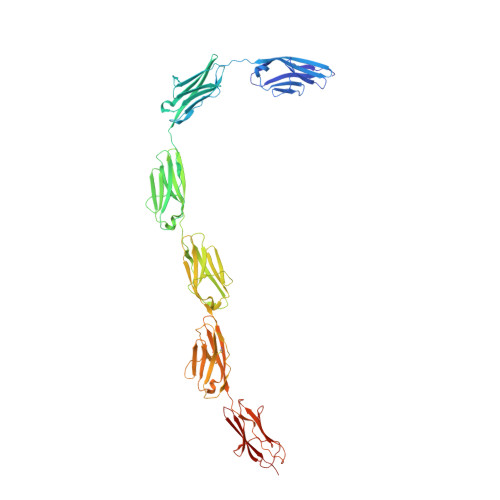A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain
von Castelmur, E., Marino, M., Svergun, D.I., Kreplak, L., Ucurum-Fotiadis, Z., Konarev, P.V., Urzhumtsev, A., Labeit, D., Labeit, S., Mayans, O.(2008) Proc Natl Acad Sci U S A 105: 1186-1191
- PubMed: 18212128
- DOI: https://doi.org/10.1073/pnas.0707163105
- Primary Citation of Related Structures:
2RIK, 2RJM, 3B43 - PubMed Abstract:
Myofibril elasticity, critical to muscle function, is dictated by the intrasarcomeric filament titin, which acts as a molecular spring. To date, the molecular events underlying the mechanics of the folded titin chain remain largely unknown. We have elucidated the crystal structure of the 6-Ig fragment I65-I70 from the elastic I-band fraction of titin and validated its conformation in solution using small angle x-ray scattering. The long-range properties of the chain have been visualized by electron microscopy on a 19-Ig fragment and modeled for the full skeletal tandem. Results show that conserved Ig-Ig transition motifs generate high-order in the structure of the filament, where conformationally stiff segments interspersed with pliant hinges form a regular pattern of dynamic super-motifs leading to segmental flexibility in the chain. Pliant hinges support molecular shape rearrangements that dominate chain behavior at moderate stretch, whereas stiffer segments predictably oppose high stretch forces upon full chain extension. There, librational entropy can be expected to act as an energy barrier to prevent Ig unfolding while, instead, triggering the unraveling of flanking springs formed by proline, glutamate, valine, and lysine (PEVK) sequences. We propose a mechanistic model based on freely jointed rigid segments that rationalizes the response to stretch of titin Ig-tandems according to molecular features.
Organizational Affiliation:
Division of Structural Biology, Biozentrum, University of Basel, Klingelbergstrasse 70, CH-4056 Basel, Switzerland.