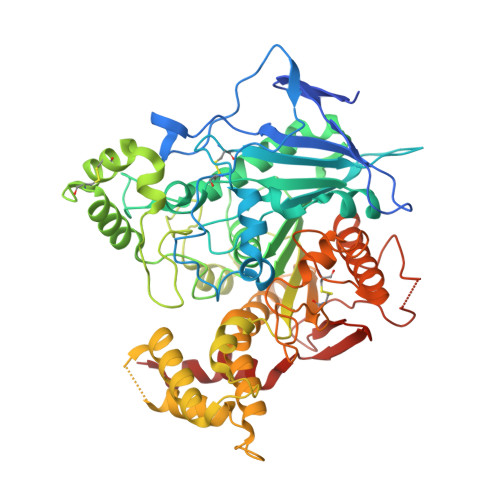
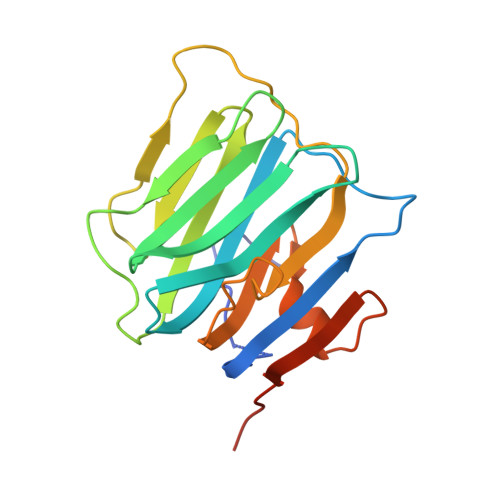
Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions.
Chen, X., Liu, H., Shim, A.H., Focia, P.J., He, X.(2008) Nat Struct Mol Biol 15: 50-56
- PubMed: 18084303
- DOI: https://doi.org/10.1038/nsmb1350
- Primary Citation of Related Structures:
3B3Q - PubMed Abstract:
The heterophilic synaptic adhesion molecules neuroligins and neurexins are essential for establishing and maintaining neuronal circuits by modulating the formation and maturation of synapses. The neuroligin-neurexin adhesion is Ca2+-dependent and regulated by alternative splicing. We report a structure of the complex at a resolution of 2.4 A between the mouse neuroligin-1 (NL1) cholinesterase-like domain and the mouse neurexin-1beta (NX1beta) LNS (laminin, neurexin and sex hormone-binding globulin-like) domain. The structure revealed a delicate neuroligin-neurexin assembly mediated by a hydrophilic, Ca2+-mediated and solvent-supplemented interface, rendering it capable of being modulated by alternative splicing and other regulatory factors. Thermodynamic data supported a mechanism wherein splicing site B of NL1 acts by modulating a salt bridge at the edge of the NL1-NX1beta interface. Mapping neuroligin mutations implicated in autism indicated that most such mutations are structurally destabilizing, supporting deficient neuroligin biosynthesis and processing as a common cause for this brain disorder.
Organizational Affiliation:
Northwestern University Feinberg School of Medicine, Department of Molecular Pharmacology & Biological Chemistry, Searle 8-417, 303 East Chicago Avenue, Chicago, Illinois 60611, USA.