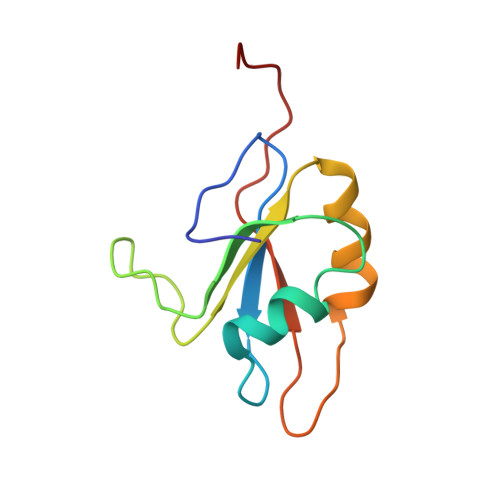
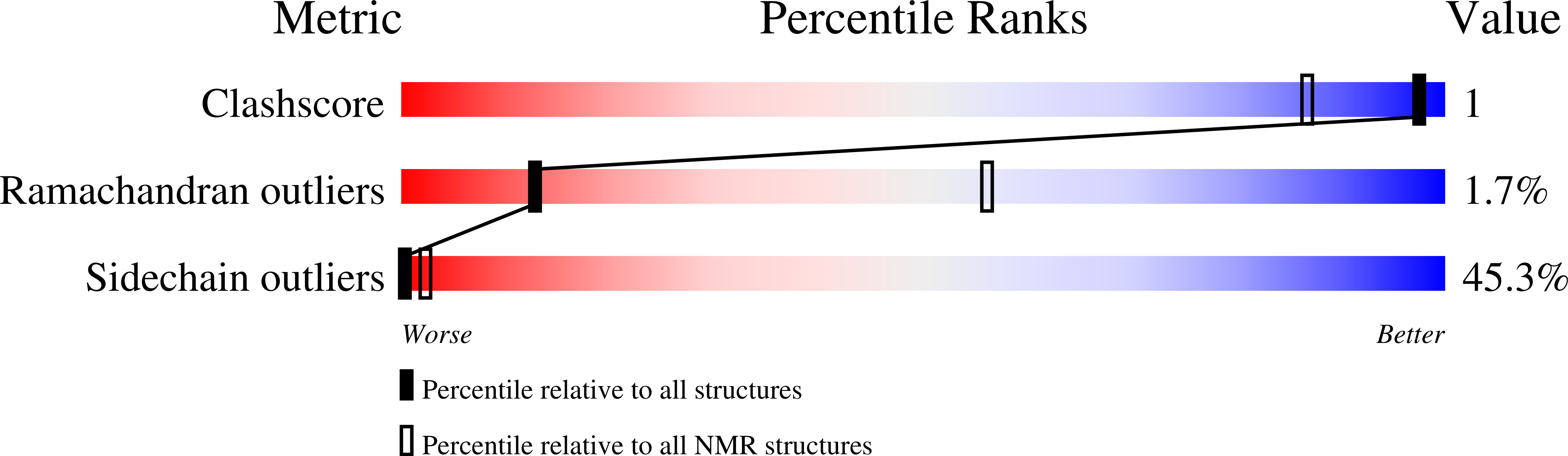Structure of an Unfolding Intermediate of an RRM Domain of ETR-3 Reveals Its Native-like Fold.
Bhatt, H., Ganguly, A.K., Sharma, S., Kushwaha, G.S., Firoz Khan, M., Sen, S., Bhavesh, N.S.(2020) Biophys J 118: 352-365
- PubMed: 31866002
- DOI: https://doi.org/10.1016/j.bpj.2019.11.3392
- Primary Citation of Related Structures:
2MY7, 2MY8 - PubMed Abstract:
Prevalence of one or more partially folded intermediates during protein unfolding with different secondary and ternary conformations has been identified as an integral character of protein unfolding. These transition-state species need to be characterized structurally for elucidation of their folding pathways. We have determined the three-dimensional structure of an intermediate state with increased conformational space sampling under urea-denaturing condition. The protein unfolds completely at 10 M urea but retains residual secondary structural propensities with restricted motion. Here, we describe the native state, observable intermediate state, and unfolded state for ETR-3 RRM-3, which has canonical RRM fold. These observations can shed more light on unfolding events for RRM-containing proteins.
Organizational Affiliation:
Transcription Regulation Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, India.