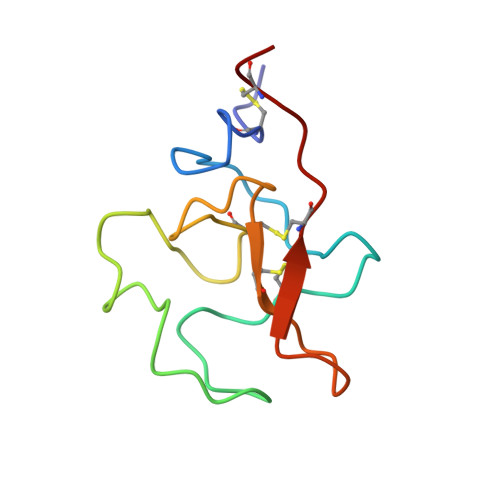
Solution structure and functional characterization of human plasminogen kringle 5.
Battistel, M.D., Grishaev, A., An, S.S., Castellino, F.J., Llinas, M.(2009) Biochemistry 48: 10208-10219
- PubMed: 19821587
- DOI: https://doi.org/10.1021/bi901433n
- Primary Citation of Related Structures:
2KNF - PubMed Abstract:
The ligand binding properties of the kringle 5 (K5) domain of human plasminogen have been investigated via intrinsic tryptophan fluorescence. The oleic acid (OA) affinity for K5 was quantified, yielding an association constant K(a) approximately 2.08 x 10(4) mM(-1). Simultaneously, it was determined that OA and trans-4-(aminomethyl)cyclohexanecarboxylic acid (AMCHA) (K(a) approximately 50 mM(-1)) compete for binding to K5. The solution structure of K5 in the presence of 11 mM AMCHA was solved via NMR spectroscopy (protein heavy atom RMSD approximately 0.93 +/- 0.12 A). The AMCHA binding site was localized via (1)H/(15)N chemical shift perturbation mapping assisted by in silico docking. We have found that AMCHA binds at the canonical kringle lysine binding site (LBS), structured by the Pro54-Gly60 segment plus the neighboring Phe36, Thr37, Trp62, Leu71, and Tyr72 residues. The segment 30-42, encompassing LBS residues, appears to be endowed with a higher degree of structural flexibility as suggested by the relatively lower value of S(2), the generalized order parameter, consistent with a higher backbone heavy atom RMSD of approximately 1.22 A (vs 0.84 A overall) between the two monomeric units in the crystal unit cell, of potential significance for ligand binding. OA was found to perturb the same area of the protein, namely, the LBS, as well as Tyr74. Combined with previous studies, the observation of OA binding expands the range of ligands that interact with kringle 5 while it widens the scope of potential biological functions for kringle domains.
Organizational Affiliation:
Department of Chemistry, Carnegie Mellon University, 4400 Fifth Avenue, Pittsburgh, Pennsylvania 15213, USA.