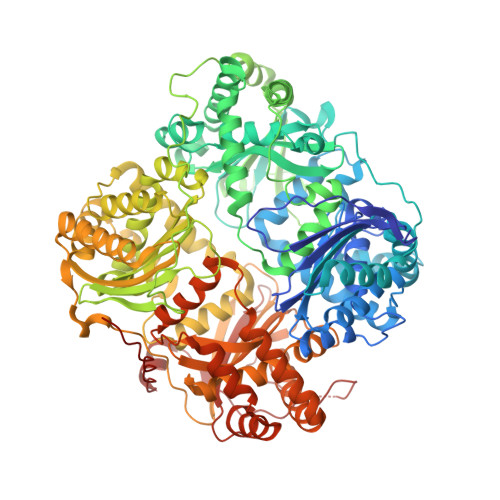
Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism.
Shen, Y., Joachimiak, A., Rosner, M.R., Tang, W.J.(2006) Nature 443: 870-874
- PubMed: 17051221
- DOI: https://doi.org/10.1038/nature05143
- Primary Citation of Related Structures:
2G47, 2G48, 2G49, 2G54, 2G56 - PubMed Abstract:
Insulin-degrading enzyme (IDE), a Zn2+-metalloprotease, is involved in the clearance of insulin and amyloid-beta (refs 1-3). Loss-of-function mutations of IDE in rodents cause glucose intolerance and cerebral accumulation of amyloid-beta, whereas enhanced IDE activity effectively reduces brain amyloid-beta (refs 4-7). Here we report structures of human IDE in complex with four substrates (insulin B chain, amyloid-beta peptide (1-40), amylin and glucagon). The amino- and carboxy-terminal domains of IDE (IDE-N and IDE-C, respectively) form an enclosed cage just large enough to encapsulate insulin. Extensive contacts between IDE-N and IDE-C keep the degradation chamber of IDE inaccessible to substrates. Repositioning of the IDE domains enables substrate access to the catalytic cavity. IDE uses size and charge distribution of the substrate-binding cavity selectively to entrap structurally diverse polypeptides. The enclosed substrate undergoes conformational changes to form beta-sheets with two discrete regions of IDE for its degradation. Consistent with this model, mutations disrupting the contacts between IDE-N and IDE-C increase IDE catalytic activity 40-fold. The molecular basis for substrate recognition and allosteric regulation of IDE could aid in designing IDE-based therapies to control cerebral amyloid-beta and blood sugar concentrations.
Organizational Affiliation:
Ben-May Institute for Cancer Research, The University of Chicago, 929 East 57th Street, Chicago, Illinois 60637, USA.