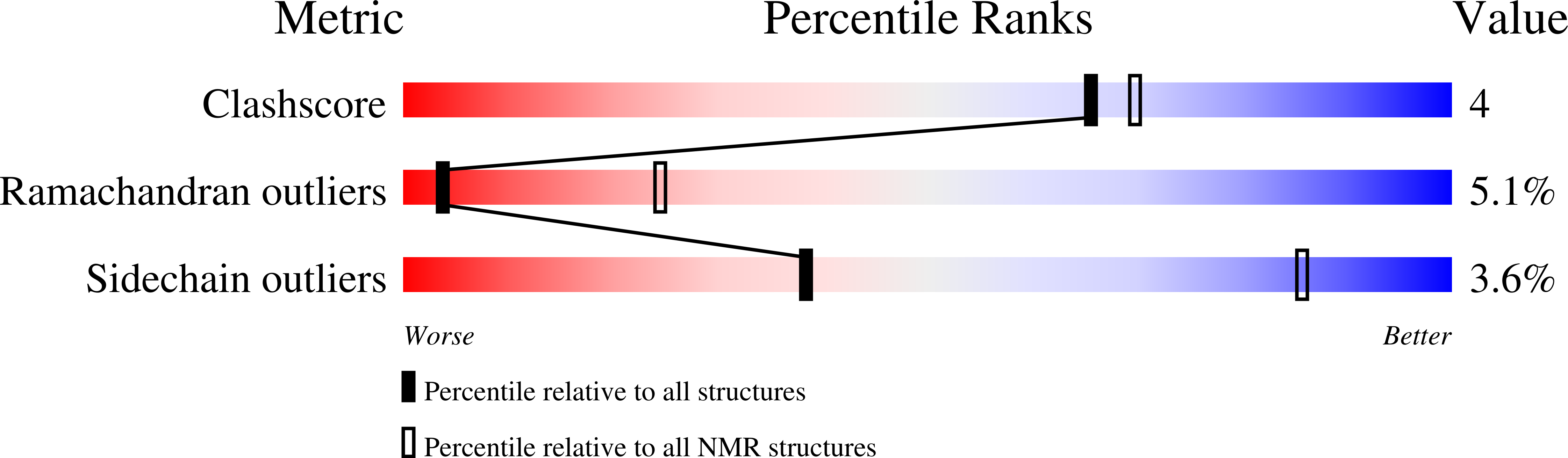The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Huth, J.R., Bewley, C.A., Nissen, M.S., Evans, J.N., Reeves, R., Gronenborn, A.M., Clore, G.M.(1997) Nat Struct Biol 4: 657-665
- PubMed: 9253416
- DOI: https://doi.org/10.1038/nsb0897-657
- Primary Citation of Related Structures:
2EZD, 2EZE, 2EZF, 2EZG - PubMed Abstract:
The solution structure of a complex between a truncated form of HMG-I(Y), consisting of the second and third DNA binding domains (residues 51-90), and a DNA dodecamer containing the PRDII site of the interferon-beta promoter has been solved by multidimensional nuclear magnetic resonance spectroscopy. The stoichiometry of the complex is one molecule of HMG-I(Y) to two molecules of DNA. The structure reveals a new architectural minor groove binding motif which stabilizes B-DNA, thereby facilitating the binding of other transcription factors in the opposing major groove. The interactions involve a central Arg-Gly-Arg motif together with two other modules that participate in extensive hydrophobic and polar contracts. The absence of one of these modules in the third DNA binding domain accounts for its-100 fold reduced affinity relative to the second one.
Organizational Affiliation:
Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892-520, USA.