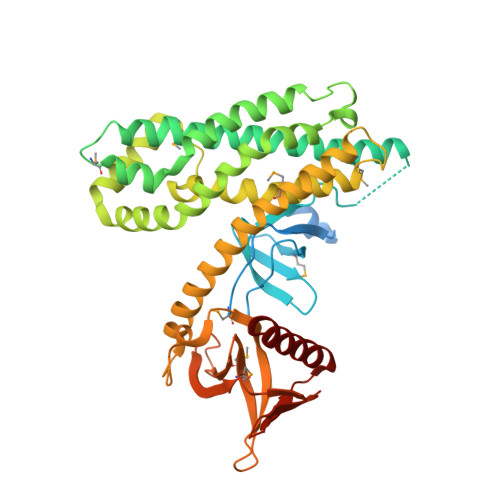
Crystal structure of the rac activator, Asef, reveals its autoinhibitory mechanism
Murayama, K., Shirouzu, M., Kawasaki, Y., Kato-Murayama, M., Hanawa-Suetsugu, K., Sakamoto, A., Katsura, Y., Suenaga, A., Toyama, M., Terada, T., Taiji, M., Akiyama, T., Yokoyama, S.(2007) J Biol Chem 282: 4238-4242
- PubMed: 17190834
- DOI: https://doi.org/10.1074/jbc.C600234200
- Primary Citation of Related Structures:
2DX1 - PubMed Abstract:
The Rac-specific guanine nucleotide exchange factor (GEF) Asef is activated by binding to the tumor suppressor adenomatous polyposis coli mutant, which is found in sporadic and familial colorectal tumors. This activated Asef is involved in the migration of colorectal tumor cells. The GEFs for Rho family GTPases contain the Dbl homology (DH) domain and the pleckstrin homology (PH) domain. When Asef is in the resting state, the GEF activity of the DH-PH module is intramolecularly inhibited by an unidentified mechanism. Asef has a Src homology 3 (SH3) domain in addition to the DH-PH module. In the present study, the three-dimensional structure of Asef was solved in its autoinhibited state. The crystal structure revealed that the SH3 domain binds intramolecularly to the DH domain, thus blocking the Rac-binding site. Furthermore, the RT-loop and the C-terminal region of the SH3 domain interact with the DH domain in a manner completely different from those for the canonical binding to a polyproline-peptide motif. These results demonstrate that the blocking of the Rac-binding site by the SH3 domain is essential for Asef autoinhibition. This may be a common mechanism in other proteins that possess an SH3 domain adjacent to a DH-PH module.
Organizational Affiliation:
Tohoku University Biomedical Engineering Research Organization, Sendai 980-8575; RIKEN Genomic Sciences Center, Yokohama Institute, Yokohama 230-0045.