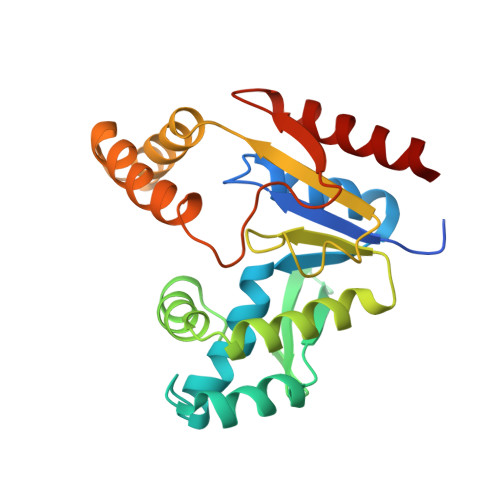
Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae
Yu, L., Mack, J., Hajduk, P.J., Kakavas, S.J., Saiki, A.Y., Lerner, C.G., Olejniczak, E.T.(2003) Protein Sci 12: 2613-2621
- PubMed: 14573872
- DOI: https://doi.org/10.1110/ps.03256803
- Primary Citation of Related Structures:
1Q3T - PubMed Abstract:
Streptococcus pneumoniae is a major human pathogen that causes high mortality and morbidity and has developed resistance to many antibiotics. We show that the gene product from SP1603, identified from S. pneumoniae TIGR4, is a CMP kinase that is essential for bacterial growth. It represents an attractive drug target for the development of a novel antibiotic to overcome the problems of drug resistance development for this organism. Here we describe the three-dimensional solution structure of the S. pneumoniae CMP kinase as determined by NMR spectroscopy. The structure consists of eight alpha-helices and two beta-sheets that fold into the classical core domain, the substrate-binding domain, and the LID domain. The three domains of the protein pack together to form a central cavity for substrate-binding and enzymatic catalysis. The S. pneumoniae CMP kinase resembles the fold of the Escherichia coli homolog. An insertion of one residue is observed at the beta-turn in the substrate-binding domain of the S. pneumoniae CMP kinase when compared with the E. coli homolog. Chemical shift perturbations caused by the binding of CMP, CDP, and ATP revealed that CMP or CDP binds to the junction between the core and substrate-binding domains, whereas ATP binds to the junction between the core and LID domains. From NMR relaxation studies, we determined that the loops in the LID domain are highly mobile. These mobile loops could aid in the closing/opening of the LID domain during enzyme catalysis.
Organizational Affiliation:
Pharmaceutical Discovery Division, Global Pharmaceutical Research and Development, Abbott Laboratories, Abbott Park, Illinois 60064-6098, USA. Liping.Yu@abbott.com