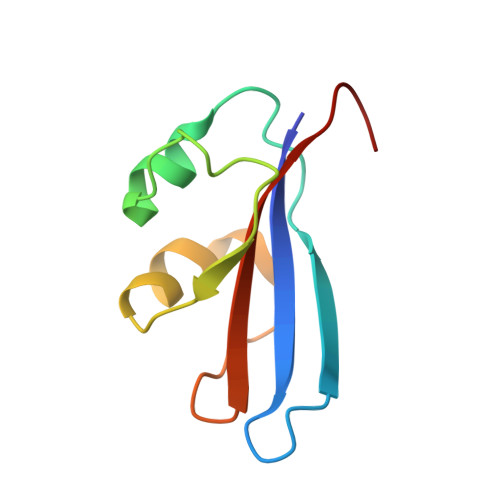
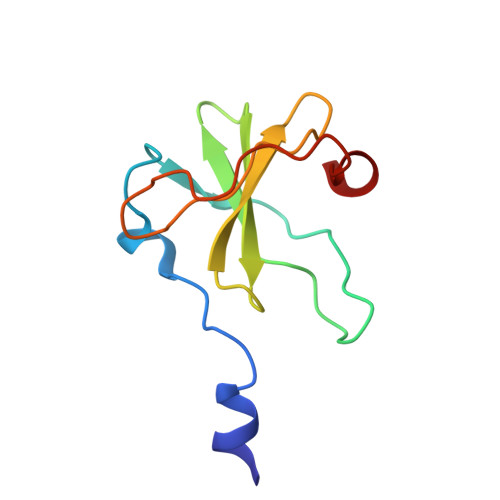
Inhibition of a Ribosome Inactivating Ribonuclease: The Crystal Structure of the Cytotoxic Domain of Colicin E3 in Complex with its Immunity Protein
Carr, S., Walker, D., James, R., Kleanthous, C., Hemmings, A.M.(2000) Structure 8: 949
- PubMed: 10986462
- DOI: https://doi.org/10.1016/s0969-2126(00)00186-6
- Primary Citation of Related Structures:
1E44 - PubMed Abstract:
The cytotoxicity of most ribonuclease E colicins towards Escherichia coli arises from their ability to specifically cleave between bases 1493 and 1494 of 16S ribosomal RNA. This activity is carried by the C-terminal domain of the colicin, an activity which if left unneutralised would lead to destruction of the producing cell. To combat this the host E. coli cell produces an inhibitor protein, the immunity protein, which forms a complex with the ribonuclease domain effectively suppressing its activity. We have solved the crystal structure of the cytotoxic domain of the ribonuclease colicin E3 in complex with its immunity protein, Im3. The structure of the ribonuclease domain, the first of its class, reveals a highly twisted central beta-sheet elaborated with a short N-terminal helix, the residues of which form a well-packed interface with the immunity protein. The structure of the ribonuclease domain of colicin E3 is novel and forms an interface with its inhibitor which is significantly different in character to that reported for the DNase colicin complexes with their immunity proteins. The structure also gives insight into the mode of action of this class of enzymatic colicins by allowing the identification of potentially catalytic residues. This in turn reveals that the inhibitor does not bind at the active site but rather at an adjacent site, leaving the catalytic centre exposed in a fashion similar to that observed for the DNase colicins. Thus, E. coli appears to have evolved similar methods for ensuring efficient inhibition of the potentially destructive effects of the two classes of enzymatic colicins.
Organizational Affiliation:
Colicin Research Group, School of Chemical Sciences, University of East Anglia, NR4 7TJ, Norwich, UK.