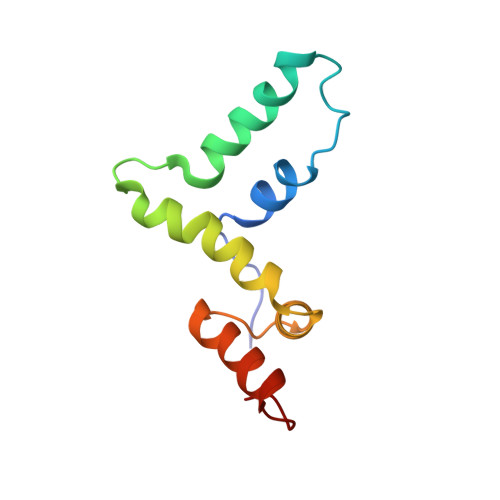
Mammalian SCAN Domain Dimer Is a Domain-Swapped Homolog of the HIV Capsid C-Terminal Domain
Ivanov, D., Stone, J.R., Maki, J.L., Collins, T., Wagner, G.(2005) Mol Cell 17: 137-143
- PubMed: 15629724
- DOI: https://doi.org/10.1016/j.molcel.2004.12.015
- Primary Citation of Related Structures:
1Y7Q - PubMed Abstract:
Retroviral assembly is driven by multiple interactions mediated by the Gag polyprotein, the main structural component of the forming viral shell. Critical determinants of Gag oligomerization are contained within the C-terminal domain (CTD) of the capsid protein, which also harbors a conserved sequence motif, the major homology region (MHR), in the otherwise highly variable Gag. An unexpected clue about the MHR function in retroviral assembly emerges from the structure of the zinc finger-associated SCAN domain we describe here. The SCAN dimer adopts a fold almost identical to that of the retroviral capsid CTD but uses an entirely different dimerization interface caused by swapping the MHR-like element between the monomers. Mutations in retroviral capsid proteins and functional data suggest that a SCAN-like MHR-swapped CTD dimer forms during immature particle assembly. In the SCAN-like dimer, the MHR contributes the major part of the large intertwined dimer interface explaining its functional significance.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.