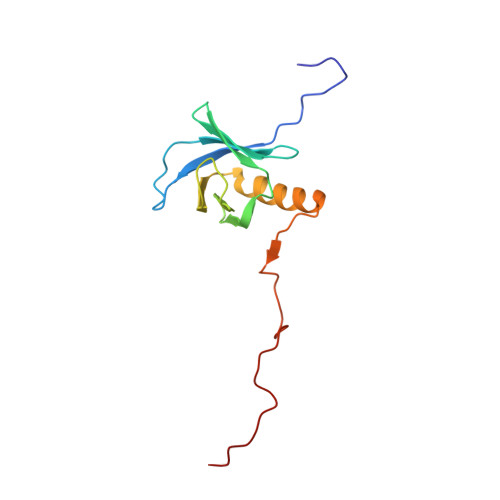
Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors
Dhalluin, C., Yan, K.S., Plotnikova, O., Lee, K.W., Zeng, L., Kuti, M., Mujtaba, S., Goldfarb, M.P., Zhou, M.-M.(2000) Mol Cell 6: 921-929
- PubMed: 11090629
- DOI: https://doi.org/10.1016/s1097-2765(05)00087-0
- Primary Citation of Related Structures:
1XR0 - PubMed Abstract:
SNT adaptor proteins transduce activation of fibroblast growth factor receptors (FGFRs) and neurotrophin receptors (TRKs) to common signaling targets. The SNT-1 phosphotyrosine binding (PTB) domain recognizes activated TRKs at a canonical NPXpY motif and, atypically, binds to nonphosphorylated FGFRs in a region lacking tyrosine or asparagine. Here, using NMR and mutational analyses, we show that the PTB domain utilizes distinct sets of amino acid residues to interact with FGFRs or TRKs in a mutually exclusive manner. The FGFR1 peptide wraps around the beta sandwich structure of the PTB domain, and its binding is possibly regulated by conformational change of a unique C-terminal beta strand in the protein. Our results suggest mechanisms by which SNTs serve as molecular switches to mediate the essential interplay between FGFR and TRK signaling during neuronal differentiation.
Organizational Affiliation:
Department of Physiology and Biophysics, Mount Sinai School of Medicine, New York University, New York 10029, USA.