Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism.
Kato, Y., Misra, S., Puertollano, R., Hurley, J.H., Bonifacino, J.S.(2002) Nat Struct Biol 9: 532-536
- PubMed: 12032548
- DOI: https://doi.org/10.1038/nsb807
- Primary Citation of Related Structures:
1LF8 - PubMed Abstract:
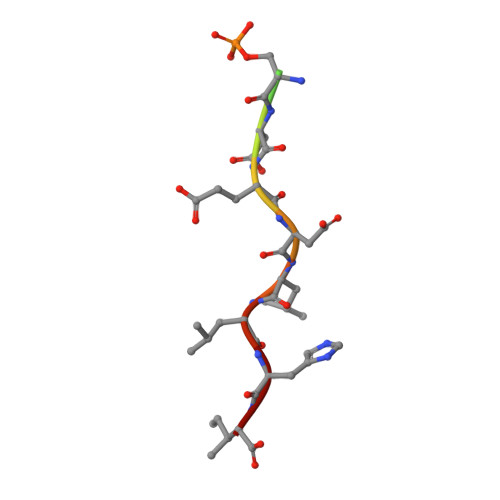
Phosphorylation of the cytosolic tails of transmembrane receptors can regulate their intracellular trafficking. The structural basis for such regulation, however, has not been explained in most cases. The cytosolic tail of the cation-independent mannose 6-phosphate receptor contains a serine residue within an acidic-cluster dileucine signal that is important for the function of the receptor in the biosynthetic sorting of lysosomal hydrolases. We show here that phosphorylation of this Ser enhances interactions of the signal with its recognition module, the VHS domain of the GGA proteins. Crystallographic analyses demonstrate that the phosphoserine residue interacts electrostatically with two basic residues on the VHS domain of GGA3, thus providing an additional point of attachment of the acidic-cluster dileucine signal to its recognition module.
Organizational Affiliation:
Cell Biology and Metabolism Branch, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland 20892, USA.