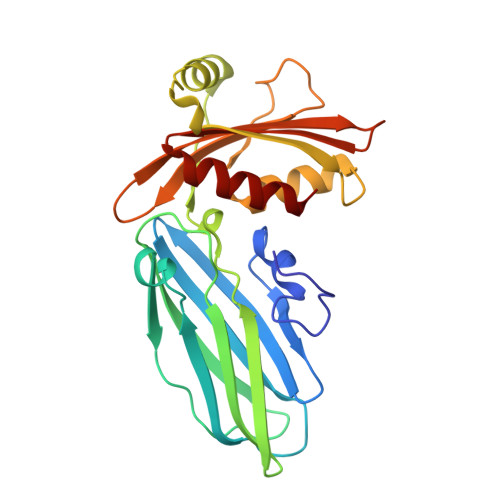
Accessory protein recruitment motifs in clathrin-mediated endocytosis.
Brett, T.J., Traub, L.M., Fremont, D.H.(2002) Structure 10: 797-809
- PubMed: 12057195
- DOI: https://doi.org/10.1016/s0969-2126(02)00784-0
- Primary Citation of Related Structures:
1KY6, 1KY7, 1KYD, 1KYF, 1KYU - PubMed Abstract:
Clathrin-mediated endocytosis depends upon the interaction of accessory proteins with the alpha-ear of the AP-2 adaptor. We present structural characterization of these regulatory interactions. DPF and DPW motif peptides derived from eps15 and epsin bind in type I beta turn conformations to a conserved pocket on the alpha-ear platform. We show evidence for a second binding site that is DPW motif specific. The structure of a complex with an AP-2 binding segment from amphiphysin reveals a novel binding motif that we term FxDxF, which is engaged in an extended conformation by a unique surface of the platform domain. The FxDxF motif is also used by AP180 and the 170 kDa isoform of synaptojanin and can be found in several potential endocytic proteins, including HIP1, CD2AP, and PLAP. A mechanism of clathrin assembly regulation is suggested by three different AP-2 engagement modes.
Organizational Affiliation:
Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO 63110, USA.