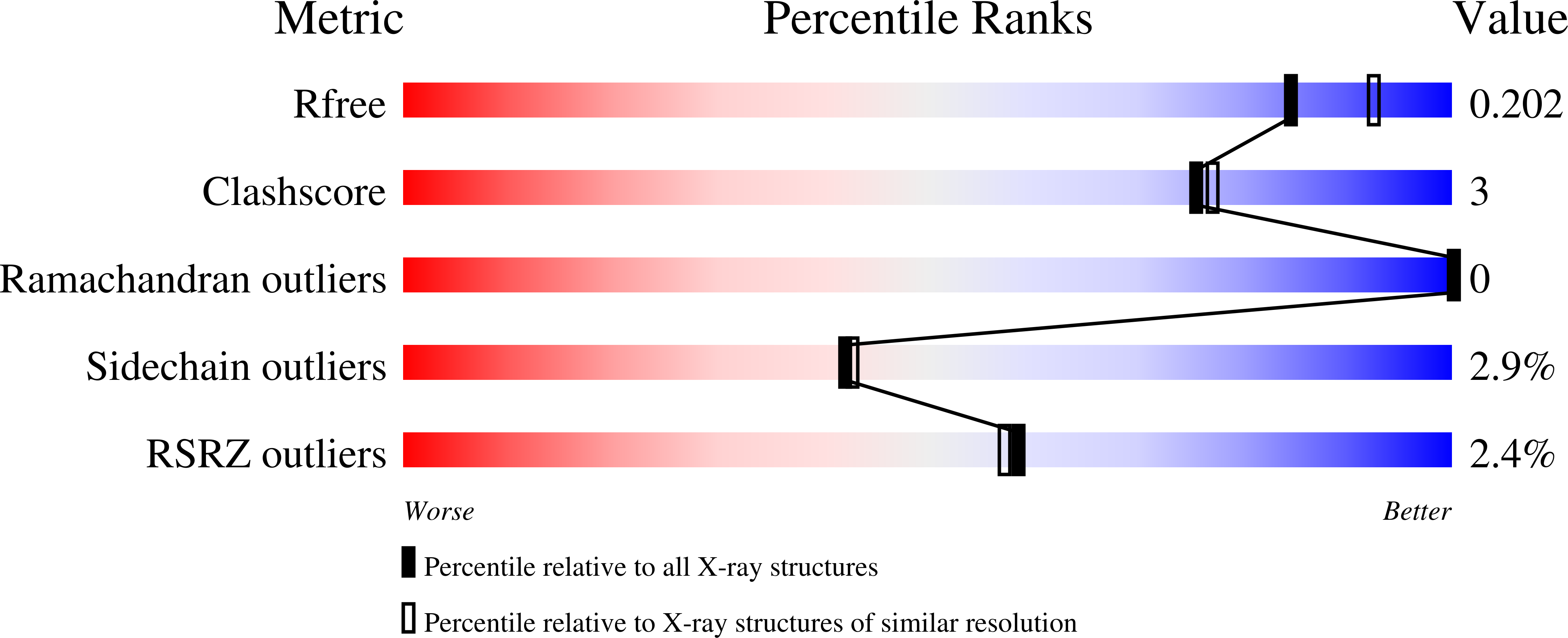
Human salivary alpha-amylase Trp58 situated at subsite -2 is critical for enzyme activity.
Ramasubbu, N., Ragunath, C., Mishra, P.J., Thomas, L.M., Kandra, L.(2004) Eur J Biochem 271: 2517-2529
- PubMed: 15182367
- DOI: https://doi.org/10.1111/j.1432-1033.2004.04182.x
- Primary Citation of Related Structures:
1JXJ, 1NM9 - PubMed Abstract:
The nonreducing end of the substrate-binding site of human salivary alpha-amylase contains two residues Trp58 and Trp59, which belong to beta2-alpha2 loop of the catalytic (beta/alpha)(8) barrel. While Trp59 stacks onto the substrate, the exact role of Trp58 is unknown. To investigate its role in enzyme activity the residue Trp58 was mutated to Ala, Leu or Tyr. Kinetic analysis of the wild-type and mutant enzymes was carried out with starch and oligosaccharides as substrates. All three mutants exhibited a reduction in specific activity (150-180-fold lower than the wild type) with starch as substrate. With oligosaccharides as substrates, a reduction in k(cat), an increase in K(m) and distinct differences in the cleavage pattern were observed for the mutants W58A and W58L compared with the wild type. Glucose was the smallest product generated by these two mutants in the hydrolysis oligosaccharides; in contrast, wild-type enzyme generated maltose as the smallest product. The production of glucose by W58L was confirmed from both reducing and nonreducing ends of CNP-labeled oligosaccharide substrates. The mutant W58L exhibited lower binding affinity at subsites -2, -3 and +2 and showed an increase in transglycosylation activity compared with the wild type. The lowered affinity at subsites -2 and -3 due to the mutation was also inferred from the electron density at these subsites in the structure of W58A in complex with acarbose-derived pseudooligosaccharide. Collectively, these results suggest that the residue Trp58 plays a critical role in substrate binding and hydrolytic activity of human salivary alpha-amylase.
Organizational Affiliation:
Department of Oral Biology, University of Medicine and Dentistry of New Jersey, Newark, NJ, USA. n.ramasubbu@umdnj.edu