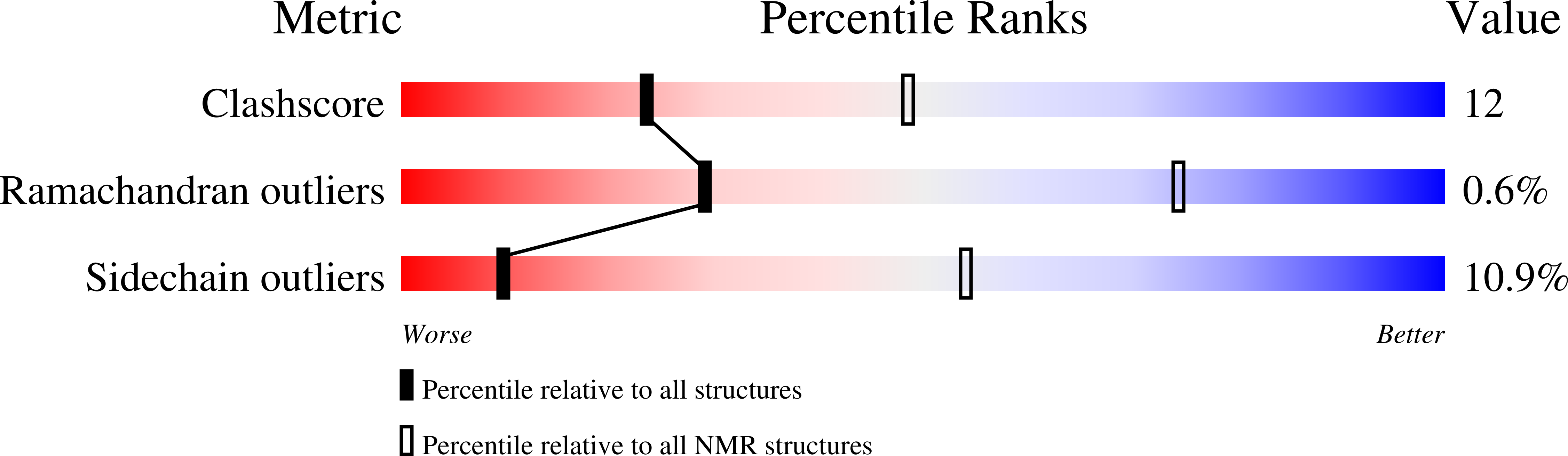
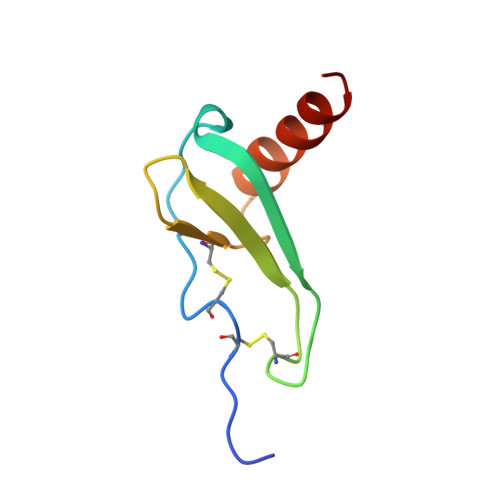
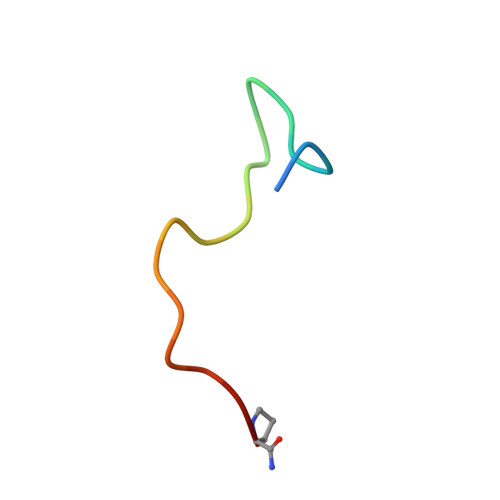
Structure of a CXC chemokine-receptor fragment in complex with interleukin-8.
Skelton, N.J., Quan, C., Reilly, D., Lowman, H.(1999) Structure 7: 157-168
- PubMed: 10368283
- DOI: https://doi.org/10.1016/S0969-2126(99)80022-7
- Primary Citation of Related Structures:
1ILP, 1ILQ - PubMed Abstract:
Interactions between CXC chemokines (e.g. interleukin-8, IL-8) and their receptors (e.g. CXCR-1) have a key role in host defense and disease by attracting and upregulating neutrophils to sites of inflammation. The transmembrane nature of the receptor impedes structure-based understanding of ligand interactions. Linear peptides based on the N-terminal, extracellular portion of the receptor CXCR-1 do bind to IL-8, however, and inhibit the binding of IL-8 to the full-length receptor. The NMR solution structure of the complex formed between IL-8 and one such receptor-based peptide indicates that a cleft between a loop and a beta hairpin constitute part of the receptor interaction surface on IL-8. Nine residues from the C terminus of the receptor peptide (corresponding to Pro21-Pro29 of CXCR-1) occupy the cleft in an extended fashion. Intermolecular contacts are mostly hydrophobic and sidechain mediated. The results offer the first details at an atomic level of the interaction between a chemokine and its receptor. Consideration of other biochemical data allow extrapolation to a model for the interaction of IL-8 with the full-length receptor. In this model, the heparin-binding residues of IL-8 are exposed, thereby allowing presentation of the chemokine from endothelial cell-surface glycosaminoglycans. This first glimpse of how IL-8 binds to its receptor provides a foundation for the structure-based design of chemokine antagonists.
Organizational Affiliation:
Department of Protein Engineering Genentech, Inc. South San Francisco, CA 94080, USA. skelly@gene.com