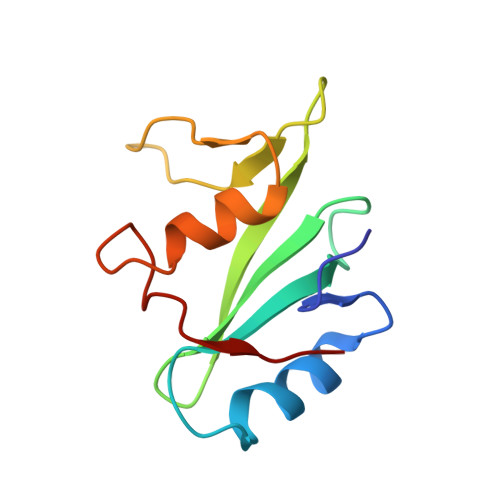
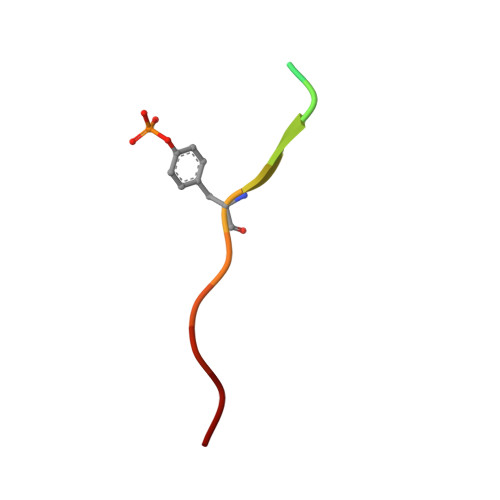
Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells.
Morra, M., Lu, J., Poy, F., Martin, M., Sayos, J., Calpe, S., Gullo, C., Howie, D., Rietdijk, S., Thompson, A., Coyle, A.J., Denny, C., Yaffe, M.B., Engel, P., Eck, M.J., Terhorst, C.(2001) Eur J Biochem 20: 5840-5852
- PubMed: 11689425
- DOI: https://doi.org/10.1093/emboj/20.21.5840
- Primary Citation of Related Structures:
1I3Z - PubMed Abstract:
The T and natural killer (NK) cell-specific gene SAP (SH2D1A) encodes a 'free SH2 domain' that binds a specific tyrosine motif in the cytoplasmic tail of SLAM (CD150) and related cell surface proteins. Mutations in SH2D1A cause the X-linked lymphoproliferative disease, a primary immunodeficiency. Here we report that a second gene encoding a free SH2 domain, EAT-2, is expressed in macrophages and B lympho cytes. The EAT-2 structure in complex with a phosphotyrosine peptide containing a sequence motif with Tyr281 of the cytoplasmic tail of CD150 is very similar to the structure of SH2D1A complexed with the same peptide. This explains the high affinity of EAT-2 for the pTyr motif in the cytoplasmic tail of CD150 but, unlike SH2D1A, EAT-2 does not bind to non-phosphorylated CD150. EAT-2 binds to the phosphorylated receptors CD84, CD150, CD229 and CD244, and acts as a natural inhibitor, which interferes with the recruitment of the tyrosine phosphatase SHP-2. We conclude that EAT-2 plays a role in controlling signal transduction through at least four receptors expressed on the surface of professional antigen-presenting cells.
Organizational Affiliation:
Division of Immunology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02115, USA. mmorra@caregroup.harvard.edu