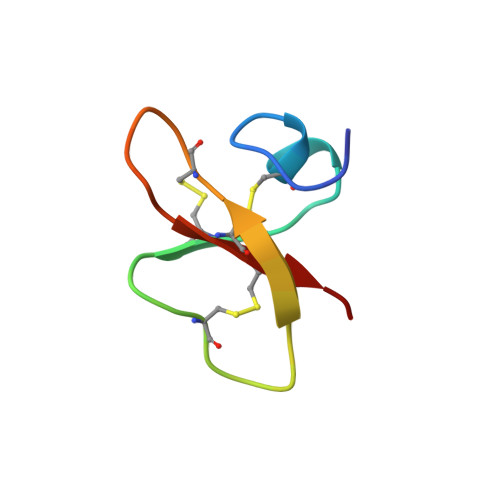
The NMR structure of human beta-defensin-2 reveals a novel alpha-helical segment.
Sawai, M.V., Jia, H.P., Liu, L., Aseyev, V., Wiencek, J.M., McCray Jr., P.B., Ganz, T., Kearney, W.R., Tack, B.F.(2001) Biochemistry 40: 3810-3816
- PubMed: 11300761
- DOI: https://doi.org/10.1021/bi002519d
- Primary Citation of Related Structures:
1FQQ - PubMed Abstract:
Human beta-defensin-2 (HBD-2) is a member of the defensin family of antimicrobial peptides. HBD-2 was first isolated from inflamed skin where it is posited to participate in the killing of invasive bacteria and in the recruitment of cells of the adaptive immune response. Static light scattering and two-dimensional proton nuclear magnetic resonance spectroscopy have been used to assess the physical state and structure of HBD-2 in solution. At concentrations of < or = 2.4 mM, HBD-2 is monomeric. The structure is amphiphilic with a nonuniform surface distribution of positive charge and contains several key structural elements, including a triple-stranded, antiparallel beta-sheet with strands 2 and 3 in a beta-hairpin conformation. A beta-bulge in the second strand occurs at Gly28, a position conserved in the entire defensin family. In solution, HBD-2 exhibits an alpha-helical segment near the N-terminus that has not been previously ascribed to solution structures of alpha-defensins or to the beta-defensin BNBD-12. This novel structural element may be a factor contributing to the specific microbicidal or chemokine-like properties of HBD-2.
Organizational Affiliation:
Departments of Microbiology and Pediatrics and NMR Facility, The University of Iowa College of Medicine, Iowa City, Iowa 52242, USA.