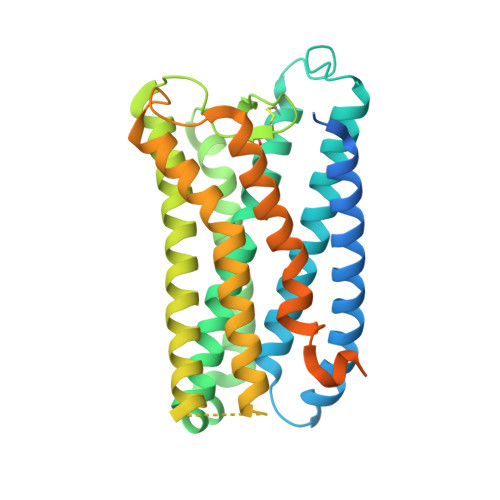
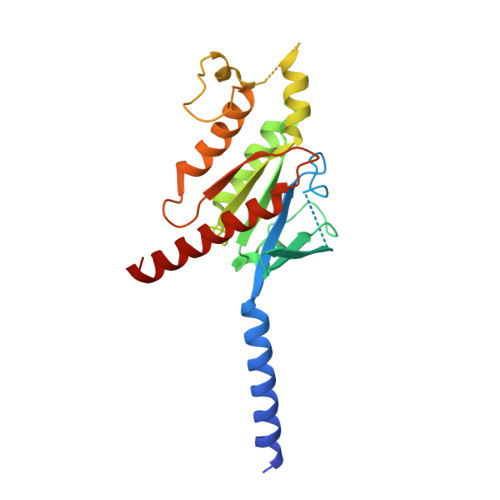
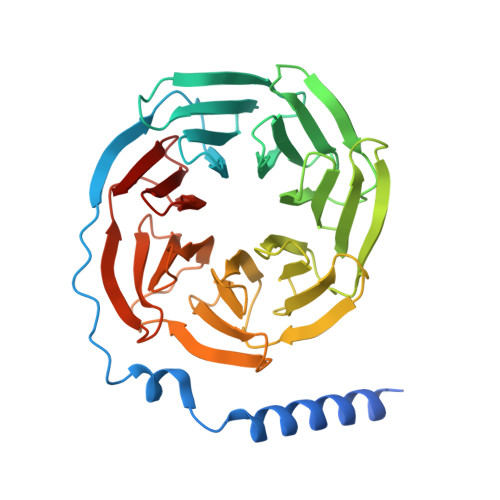
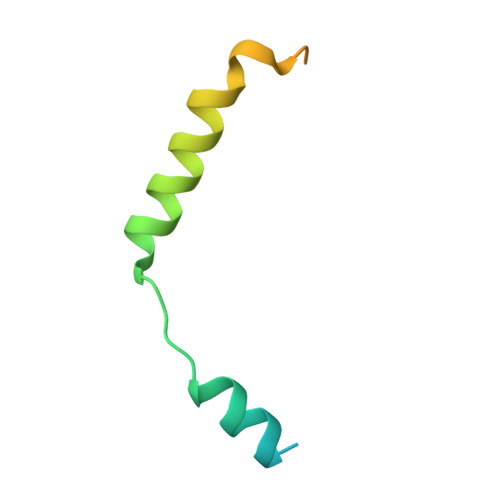
Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Lv, X., Gao, K., Nie, J., Zhang, X., Zhang, S., Ren, Y., Sun, X., Li, Q., Huang, J., Liu, L., Zhang, X., Zhang, W., Liu, X.(2023) Nat Commun 14: 8136-8136
- PubMed: 38065938
- DOI: https://doi.org/10.1038/s41467-023-43922-8
- Primary Citation of Related Structures:
8IQ4, 8IQ6 - PubMed Abstract:
Prostaglandins and their receptors regulate various physiological processes. Carboprost, an analog of prostaglandin F 2α and an agonist for the prostaglandin F2-alpha receptor (FP receptor), is clinically used to treat postpartum hemorrhage (PPH). However, off-target activation of closely related receptors such as the prostaglandin E receptor subtype EP3 (EP3 receptor) by carboprost results in side effects and limits the clinical application. Meanwhile, the FP receptor selective agonist latanoprost is not suitable to treat PPH due to its poor solubility and fast clearance. Here, we present two cryo-EM structures of the FP receptor bound to carboprost and latanoprost-FA (the free acid form of latanoprost) at 2.7 Å and 3.2 Å resolution, respectively. The structures reveal the molecular mechanism of FP receptor selectivity for both endogenous prostaglandins and clinical drugs, as well as the molecular mechanism of G protein coupling preference by the prostaglandin receptors. The structural information may guide the development of better prostaglandin drugs.
Organizational Affiliation:
Department of Obstetrics, Xiangya Hospital Central South University, Changsha, China.