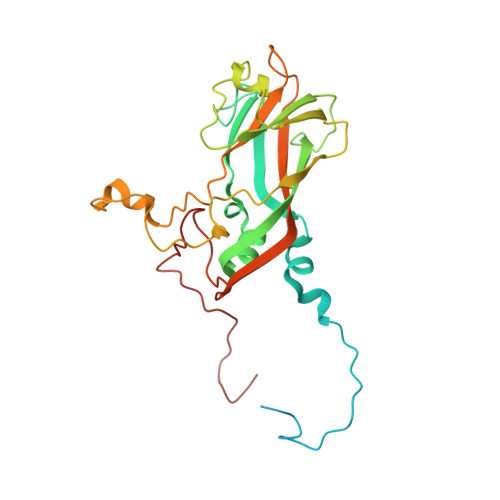
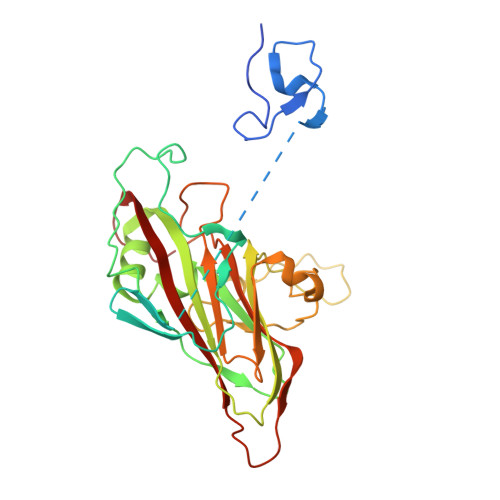
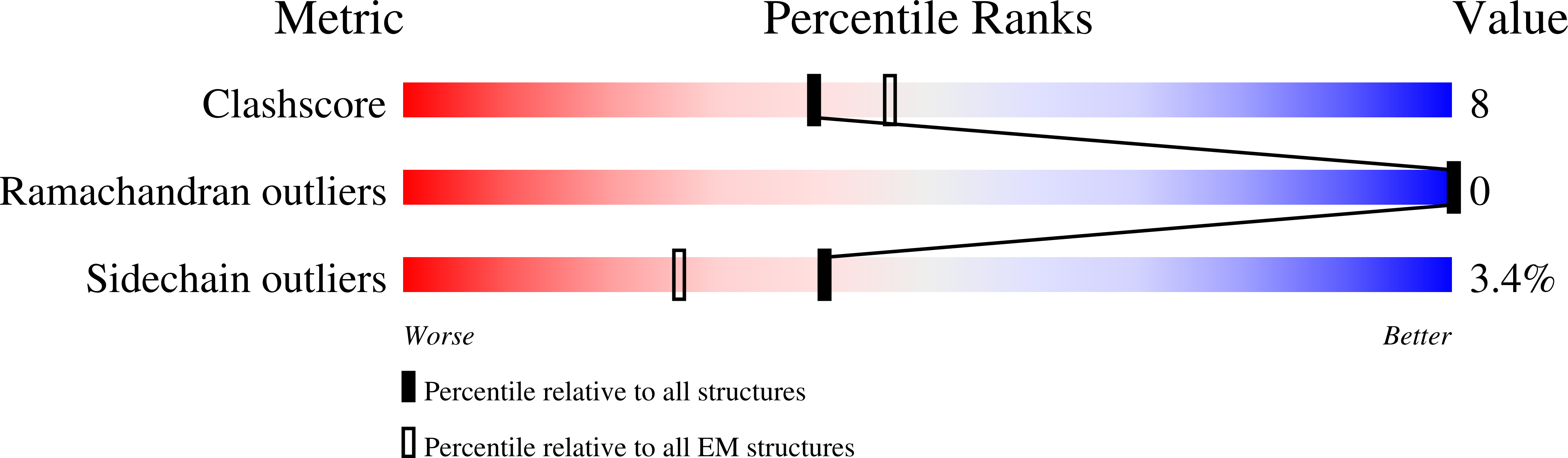Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate.
Kingston, N.J., Snowden, J.S., Martyna, A., Shegdar, M., Grehan, K., Tedcastle, A., Pegg, E., Fox, H., Macadam, A.J., Martin, J., Hogle, J.M., Rowlands, D.J., Stonehouse, N.J.(2023) bioRxiv
- PubMed: 36778240
- DOI: https://doi.org/10.1101/2023.01.30.526315
- Primary Citation of Related Structures:
8C6D - PubMed Abstract:
Enterovirus A71 (EVA71) causes widespread disease in young children with occasional fatal consequences. In common with other picornaviruses, both empty capsids (ECs) and infectious virions are produced during the viral lifecycle. While initially antigenically indistinguishable from virions, ECs readily convert to an expanded conformation at moderate temperatures. In the closely related poliovirus, these conformational changes result in loss of antigenic sites required to elicit protective immune responses. Whether this is true for EVA71 remains to be determined and is the subject of this investigation. We previously reported the selection of a thermally resistant EVA71 genogroup B2 population using successive rounds of heating and passage. The mutations found in the structural protein-coding region of the selected population conferred increased thermal stability to both virions and naturally produced ECs. Here, we introduced these mutations into a recombinant expression system to produce stabilised virus-like particles (VLPs) in Pichia pastoris . The stabilised VLPs retain the native virion-like antigenic conformation as determined by reactivity with a specific antibody. Structural studies suggest multiple potential mechanisms of antigenic stabilisation, however, unlike poliovirus, both native and expanded EVA71 particles elicited antibodies able to directly neutralise virus in vitro . Therefore, the anti-EVA71 neutralising antibodies are elicited by sites which are not canonically associated with the native conformation, but whether antigenic sites specific to the native conformation provide additional protective responses in vivo remains unclear. VLPs are likely to provide cheaper and safer alternatives for vaccine production and these data show that VLP vaccines are comparable with inactivated virus vaccines at inducing neutralising antibodies.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology, School of Molecular & Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, United Kingdom.