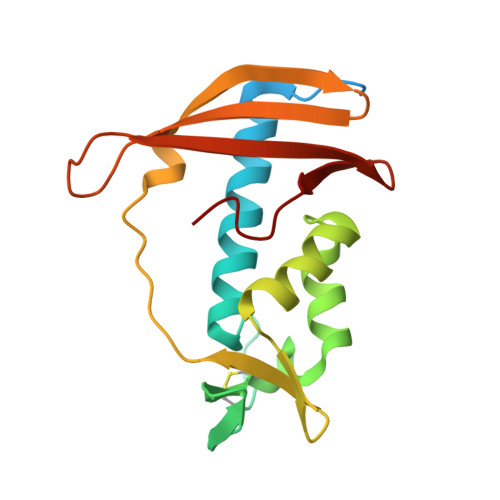
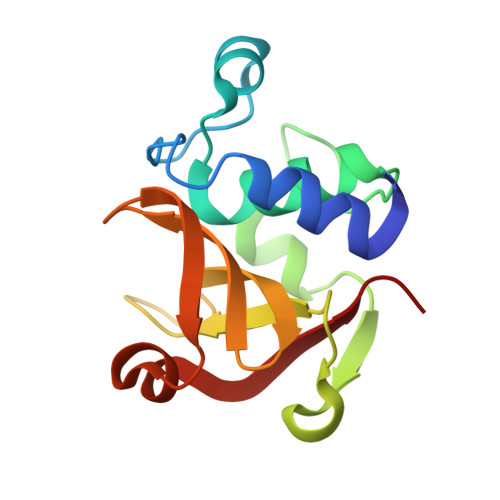
Structural analysis of the peptidoglycan DL-endopeptidase CwlO complexed with its inhibitory protein IseA.
Tandukar, S., Kwon, E., Kim, D.Y.(2024) FEBS J 291: 3723-3736
- PubMed: 38840475
- DOI: https://doi.org/10.1111/febs.17197
- Primary Citation of Related Structures:
8WT3, 8WT4 - PubMed Abstract:
Peptidoglycan DL-endopeptidases locally cleave the peptide stem of peptidoglycan in the bacterial cell wall. This process facilitates bacterial growth and division by loosening the rigid peptidoglycan layer. IseA binds to the active site of multiple DL-endopeptidases and inhibits excessive peptidoglycan degradation that leads to cell lysis. To better understand how IseA inhibits DL-endopeptidase activity, we determined the crystal structure of the peptidoglycan DL-endopeptidase CwlO/IseA complex and compared it with that of the peptidoglycan DL-endopeptidase LytE/IseA complex. Structural analyses showed significant differences between the hydrophobic pocket-binding residues of the DL-endopeptidases (F361 of CwlO and W237 of LytE). Additionally, binding assays showed that the F361 mutation of CwlO to the bulkier hydrophobic residue, tryptophan, increased its binding affinity for IseA, whereas mutation to alanine reduced the affinity. These analyses revealed that the hydrophobic pocket-binding residue of DL-endopeptidases determines IseA-binding affinity and is required for substrate-mimetic inhibition by IseA.
Organizational Affiliation:
College of Pharmacy, Yeungnam University, Gyeongsan, Korea.