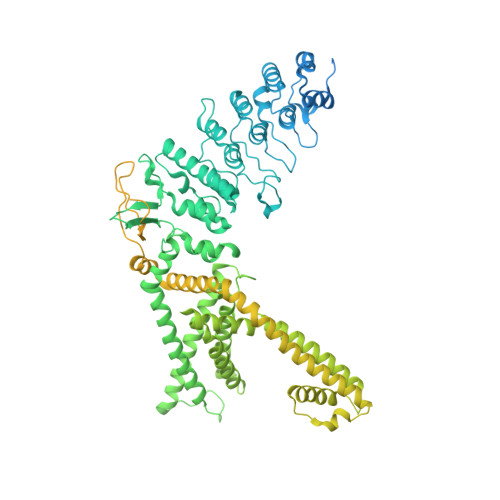
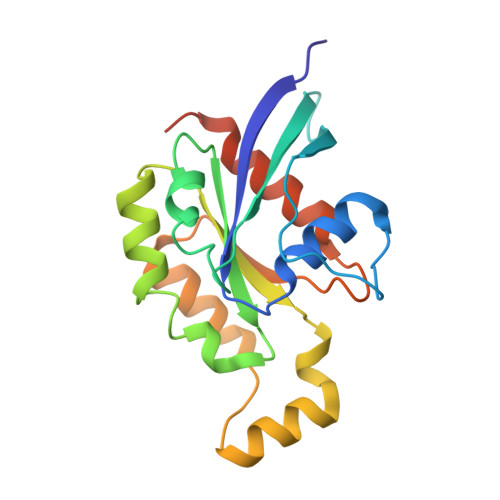
Structural Pharmacology of TRPV4 Antagonists.
Fan, J., Guo, C., Liao, D., Ke, H., Lei, J., Xie, W., Tang, Y., Tominaga, M., Huang, Z., Lei, X.(2024) Adv Sci (Weinh) 11: e2401583-e2401583
- PubMed: 38659239
- DOI: https://doi.org/10.1002/advs.202401583
- Primary Citation of Related Structures:
8JU5, 8JU6, 8JVI, 8JVJ - PubMed Abstract:
The nonselective calcium-permeable Transient Receptor Potential Cation Channel Subfamily V Member4 (TRPV4) channel regulates various physiological activities. Dysfunction of TRPV4 is linked to many severe diseases, including edema, pain, gastrointestinal disorders, lung diseases, and inherited neurodegeneration. Emerging TRPV4 antagonists show potential clinical benefits. However, the molecular mechanisms of TRPV4 antagonism remain poorly understood. Here, cryo-electron microscopy (cryo-EM) structures of human TRPV4 are presented in-complex with two potent antagonists, revealing the detailed binding pockets and regulatory mechanisms of TRPV4 gating. Both antagonists bind to the voltage-sensing-like domain (VSLD) and stabilize the channel in closed states. These two antagonists induce TRPV4 to undergo an apparent fourfold to twofold symmetry transition. Moreover, it is demonstrated that one of the antagonists binds to the VSLD extended pocket, which differs from the canonical VSLD pocket. Complemented with functional and molecular dynamics simulation results, this study provides crucial mechanistic insights into TRPV4 regulation by small-molecule antagonists, which may facilitate future drug discovery targeting TRPV4.
Organizational Affiliation:
Beijing National Laboratory for Molecular Sciences, Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, College of Chemistry and Molecular Engineering, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing, 100871, China.