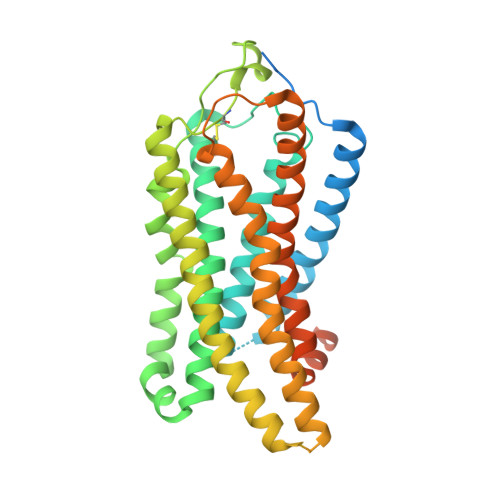
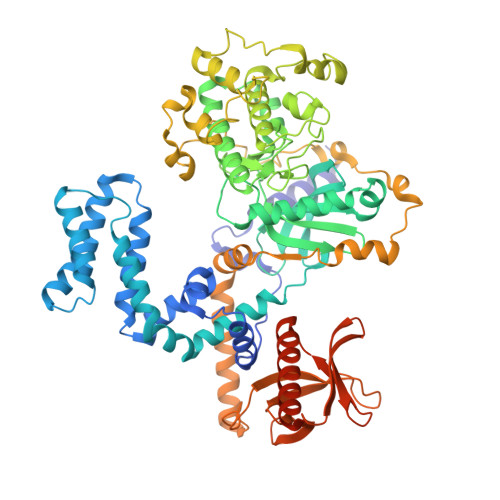
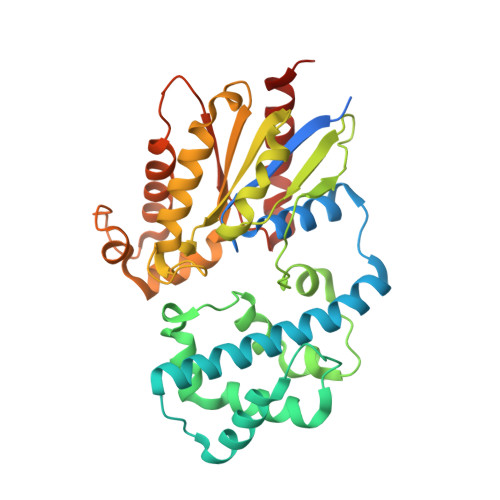
GPCR activation and GRK2 assembly by a biased intracellular agonist.
Duan, J., Liu, H., Zhao, F., Yuan, Q., Ji, Y., Cai, X., He, X., Li, X., Li, J., Wu, K., Gao, T., Zhu, S., Lin, S., Wang, M.W., Cheng, X., Yin, W., Jiang, Y., Yang, D., Xu, H.E.(2023) Nature 620: 676-681
- PubMed: 37532940
- DOI: https://doi.org/10.1038/s41586-023-06395-9
- Primary Citation of Related Structures:
8JPB, 8JPC, 8JPD, 8JPE, 8JPF - PubMed Abstract:
Phosphorylation of G-protein-coupled receptors (GPCRs) by GPCR kinases (GRKs) desensitizes G-protein signalling and promotes arrestin signalling, which is also modulated by biased ligands 1-6 . The molecular assembly of GRKs on GPCRs and the basis of GRK-mediated biased signalling remain largely unknown owing to the weak GPCR-GRK interactions. Here we report the complex structure of neurotensin receptor 1 (NTSR1) bound to GRK2, Gα q and the arrestin-biased ligand SBI-553 7 . The density map reveals the arrangement of the intact GRK2 with the receptor, with the N-terminal helix of GRK2 docking into the open cytoplasmic pocket formed by the outward movement of the receptor transmembrane helix 6, analogous to the binding of the G protein to the receptor. SBI-553 binds at the interface between GRK2 and NTSR1 to enhance GRK2 binding. The binding mode of SBI-553 is compatible with arrestin binding but clashes with the binding of Gα q protein, thus providing a mechanism for its arrestin-biased signalling capability. In sum, our structure provides a rational model for understanding the details of GPCR-GRK interactions and GRK2-mediated biased signalling.
Organizational Affiliation:
State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China. duanjia@simm.ac.cn.