Structure-guided engineering enables E3 ligase-free and versatile protein ubiquitination via UBE2E1.
Wu, X., Du, Y., Liang, L.J., Ding, R., Zhang, T., Cai, H., Tian, X., Pan, M., Liu, L.(2024) Nat Commun 15: 1266-1266
- PubMed: 38341401
- DOI: https://doi.org/10.1038/s41467-024-45635-y
- Primary Citation of Related Structures:
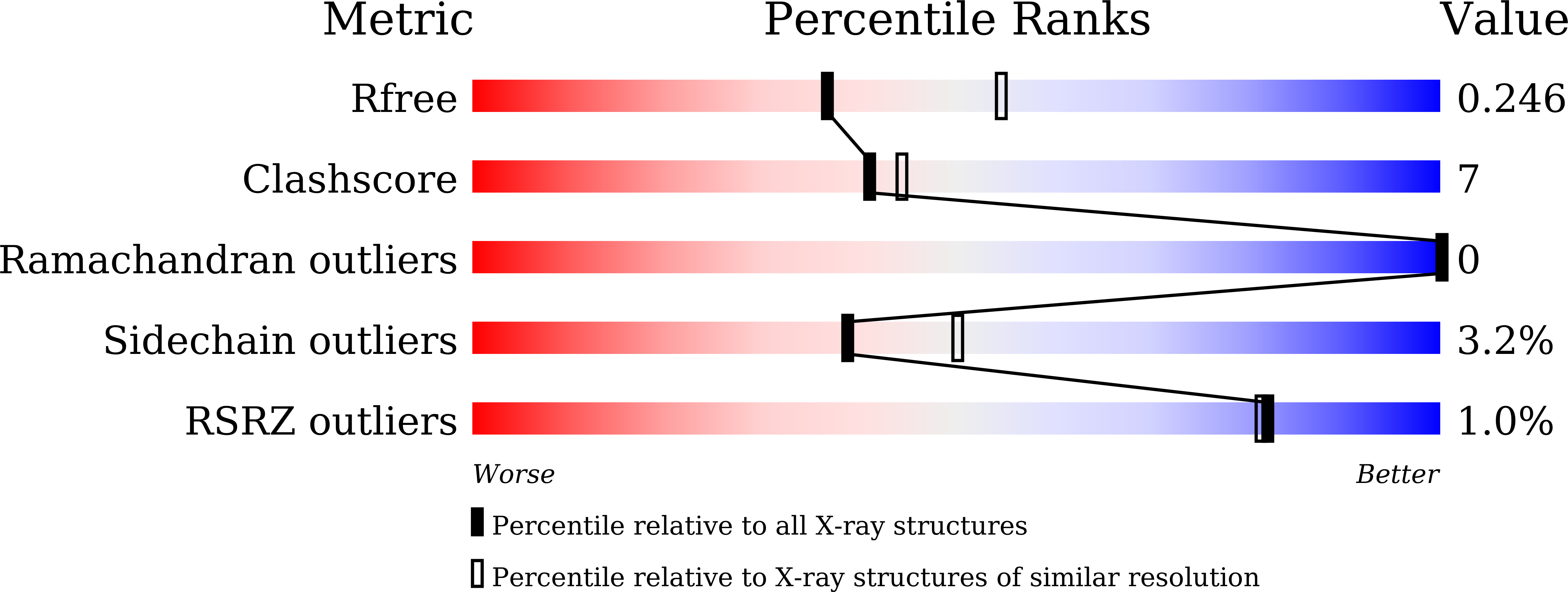
8IYA - PubMed Abstract:
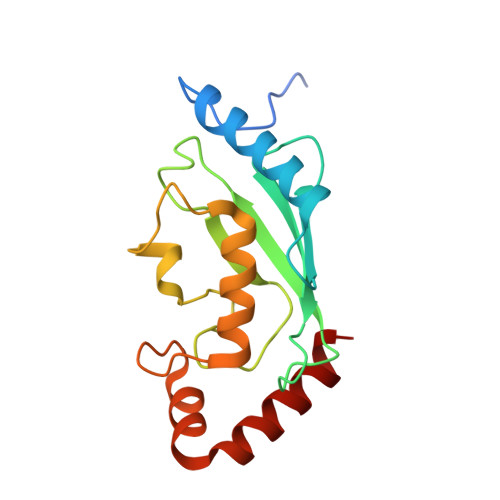
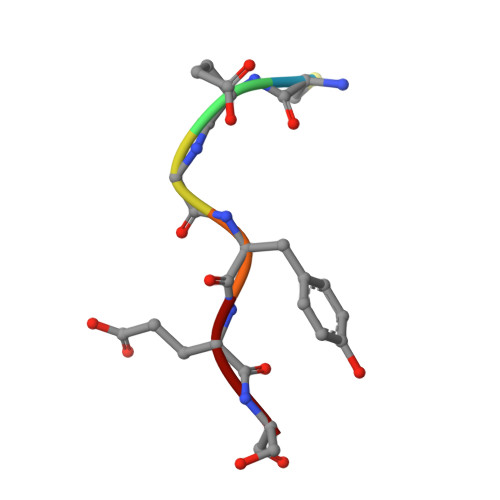
Ubiquitination, catalyzed usually by a three-enzyme cascade (E1, E2, E3), regulates various eukaryotic cellular processes. E3 ligases are the most critical components of this catalytic cascade, determining both substrate specificity and polyubiquitination linkage specificity. Here, we reveal the mechanism of a naturally occurring E3-independent ubiquitination reaction of a unique human E2 enzyme UBE2E1 by solving the structure of UBE2E1 in complex with substrate SETDB1-derived peptide. Guided by this peptide sequence-dependent ubiquitination mechanism, we developed an E3-free enzymatic strategy SUE1 (sequence-dependent ubiquitination using UBE2E1) to efficiently generate ubiquitinated proteins with customized ubiquitinated sites, ubiquitin chain linkages and lengths. Notably, this strategy can also be used to generate site-specific branched ubiquitin chains or even NEDD8-modified proteins. Our work not only deepens the understanding of how an E3-free substrate ubiquitination reaction occurs in human cells, but also provides a practical approach for obtaining ubiquitinated proteins to dissect the biochemical functions of ubiquitination.
Organizational Affiliation:
New Cornerstone Science Laboratory, Tsinghua-Peking Joint Center for Life Sciences, MOE Key Laboratory of Bioorganic Phosphorus Chemistry and Chemical Biology, Center for Synthetic and Systems Biology, Department of Chemistry, Tsinghua University, Beijing, 100084, China.