Structure-Activity Relationship of a Pyrrole Based Series of PfPKG Inhibitors as Anti-Malarials.
Gilleran, J.A., Ashraf, K., Delvillar, M., Eck, T., Fondekar, R., Miller, E.B., Hutchinson, A., Dong, A., Seitova, A., De Souza, M.L., Augeri, D., Halabelian, L., Siekierka, J., Rotella, D.P., Gordon, J., Childers, W.E., Grier, M.C., Staker, B.L., Roberge, J.Y., Bhanot, P.(2024) J Med Chem 67: 3467-3503
- PubMed: 38372781
- DOI: https://doi.org/10.1021/acs.jmedchem.3c01795
- Primary Citation of Related Structures:
8EM8 - PubMed Abstract:
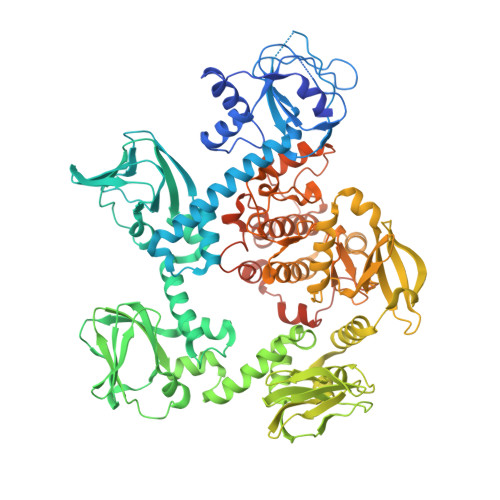
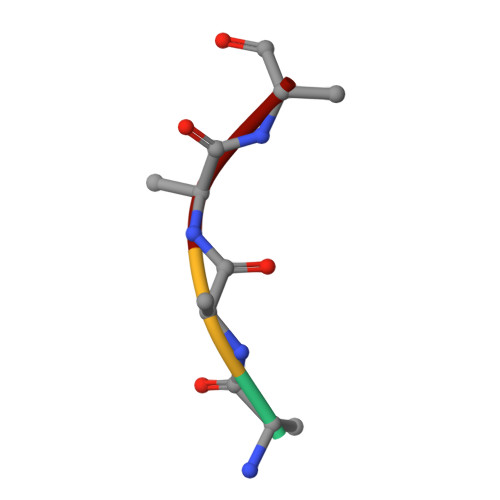
Controlling malaria requires new drugs against Plasmodium falciparum . The P. falciparum cGMP-dependent protein kinase (PfPKG) is a validated target whose inhibitors could block multiple steps of the parasite's life cycle. We defined the structure-activity relationship (SAR) of a pyrrole series for PfPKG inhibition. Key pharmacophores were modified to enable full exploration of chemical diversity and to gain knowledge about an ideal core scaffold. In vitro potency against recombinant PfPKG and human PKG were used to determine compound selectivity for the parasite enzyme. P. berghei sporozoites and P. falciparum asexual blood stages were used to assay multistage antiparasitic activity. Cellular specificity of compounds was evaluated using transgenic parasites expressing PfPKG carrying a substituted "gatekeeper" residue. The structure of PfPKG bound to an inhibitor was solved, and modeling using this structure together with computational tools was utilized to understand SAR and establish a rational strategy for subsequent lead optimization.
Organizational Affiliation:
Rutgers Molecular Design and Synthesis Core, Office for Research, Rutgers University, 610 Taylor Road, Piscataway, New Jersey 08854, United States.