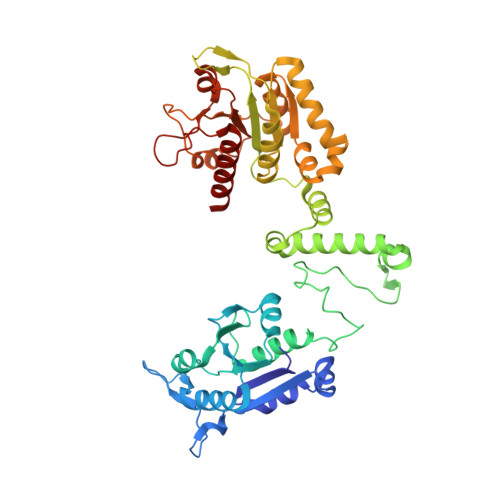
Are there double knots in proteins? Prediction and in vitro verification based on TrmD-Tm1570 fusion from C. nitroreducens.
Perlinska, A.P., Nguyen, M.L., Pilla, S.P., Staszor, E., Lewandowska, I., Bernat, A., Purta, E., Augustyniak, R., Bujnicki, J.M., Sulkowska, J.I.(2023) Front Mol Biosci 10: 1223830-1223830
- PubMed: 38903539
- DOI: https://doi.org/10.3389/fmolb.2023.1223830
- Primary Citation of Related Structures:
8B1N - PubMed Abstract:
We have been aware of the existence of knotted proteins for over 30 years-but it is hard to predict what is the most complicated knot that can be formed in proteins. Here, we show new and the most complex knotted topologies recorded to date-double trefoil knots (3 1 # 3 1 ). We found five domain arrangements (architectures) that result in a doubly knotted structure in almost a thousand proteins. The double knot topology is found in knotted membrane proteins from the CaCA family, that function as ion transporters, in the group of carbonic anhydrases that catalyze the hydration of carbon dioxide, and in the proteins from the SPOUT superfamily that gathers 3 1 knotted methyltransferases with the active site-forming knot. For each family, we predict the presence of a double knot using AlphaFold and RoseTTaFold structure prediction. In the case of the TrmD-Tm1570 protein, which is a member of SPOUT superfamily, we show that it folds in vitro and is biologically active. Our results show that this protein forms a homodimeric structure and retains the ability to modify tRNA, which is the function of the single-domain TrmD protein. However, how the protein folds and is degraded remains unknown.
Organizational Affiliation:
Centre of New Technologies, University of Warsaw, Warsaw, Poland.