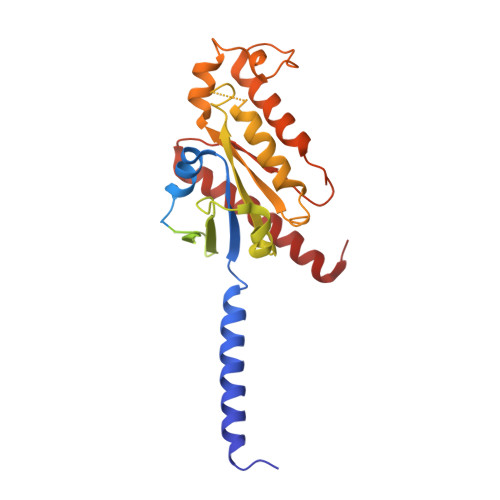
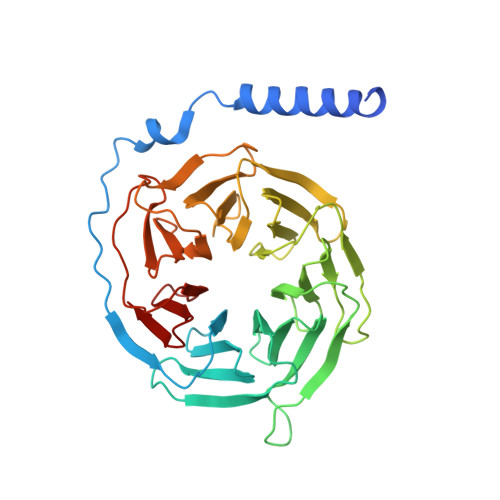
Structural basis of the ligand binding and signaling mechanism of melatonin receptors.
Wang, Q., Lu, Q., Guo, Q., Teng, M., Gong, Q., Li, X., Du, Y., Liu, Z., Tao, Y.(2022) Nat Commun 13: 454-454
- PubMed: 35075127
- DOI: https://doi.org/10.1038/s41467-022-28111-3
- Primary Citation of Related Structures:
7VGY, 7VGZ, 7VH0 - PubMed Abstract:
Melatonin receptors (MT 1 and MT 2 in humans) are family A G protein-coupled receptors that respond to the neurohormone melatonin to regulate circadian rhythm and sleep. Numerous efforts have been made to develop drugs targeting melatonin receptors for the treatment of insomnia, circadian rhythm disorder, and cancer. However, designing subtype-selective melatonergic drugs remains challenging. Here, we report the cryo-EM structures of the MT 1 -G i signaling complex with 2-iodomelatonin and ramelteon and the MT 2 -G i signaling complex with ramelteon. These structures, together with the reported functional data, reveal that although MT 1 and MT 2 possess highly similar orthosteric ligand-binding pockets, they also display distinctive features that could be targeted to design subtype-selective drugs. The unique structural motifs in MT 1 and MT 2 mediate structural rearrangements with a particularly wide opening on the cytoplasmic side. G i is engaged in the receptor core shared by MT 1 and MT 2 and presents a conformation deviating from those in other G i complexes. Together, our results provide new clues for designing melatonergic drugs and further insights into understanding the G protein coupling mechanism.
Organizational Affiliation:
Department of Clinical Laboratory, The First Affiliated Hospital of USTC, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Biomedical Sciences and Health Laboratory of Anhui Province, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, 230027, Hefei, P.R. China.