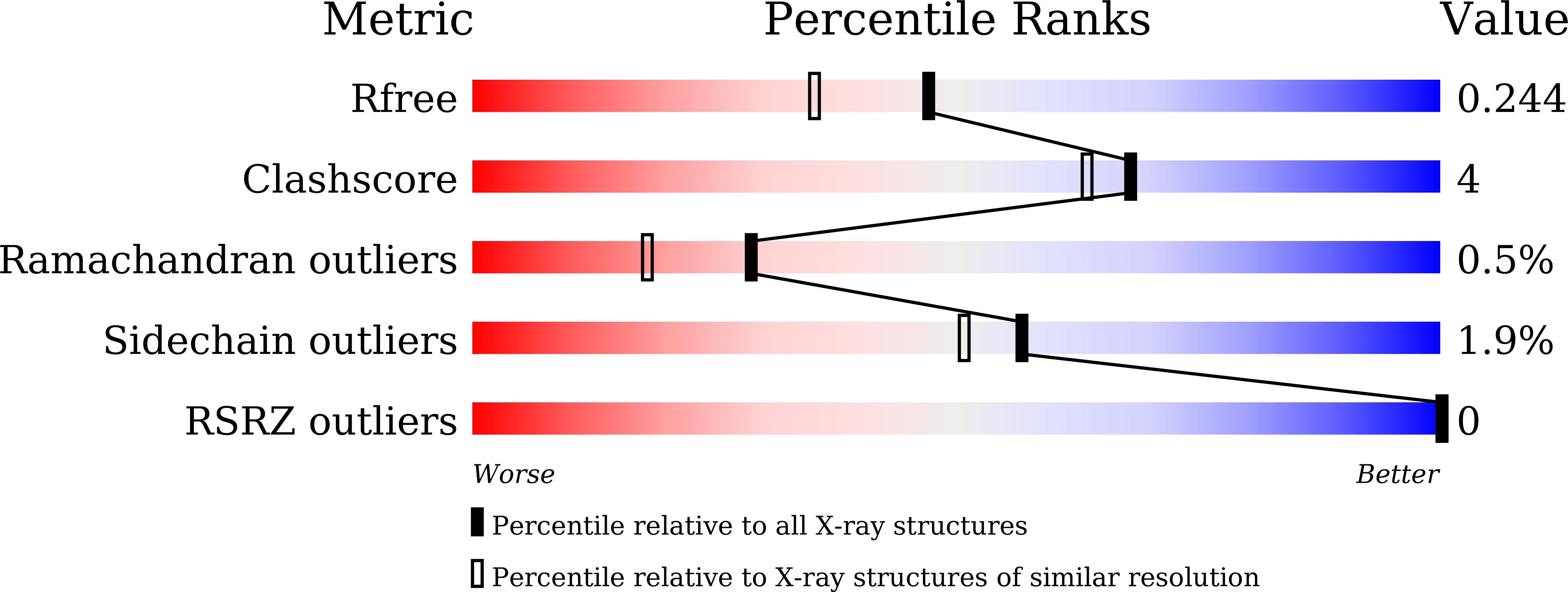
Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Yu, Y., Guo, J., Cai, Z., Ju, Y., Xu, J., Gu, Q., Zhou, H.(2021) Bioorg Chem 114: 105040-105040
- PubMed: 34098257
- DOI: https://doi.org/10.1016/j.bioorg.2021.105040
- Primary Citation of Related Structures:
7DOR, 7DPR, 7DPS, 7DQF, 7DQH, 7DQI, 7DQJ, 7DQL, 7DQM, 7DQS, 7DQU, 7DQW - PubMed Abstract:
DNA gyrase is an essential DNA topoisomerase that exists only in bacteria. Since novobiocin was withdrawn from the market, new scaffolds and new mechanistic GyrB inhibitors are urgently needed. In this study, we employed fragment screening and X-ray crystallography to identify new building blocks, as well as their binding mechanisms, to support the discovery of new GyrB inhibitors. In total, 84 of the 618 chemical fragments were shown to either thermally stabilize the ATPase domain of Escherichia coli GyrB or inhibit the ATPase activity of E. coli gyrase. Among them, the IC 50 values of fragments 10 and 23 were determined to be 605.3 μM and 446.2 μM, respectively. Cocrystal structures of the GyrB ATPase domain with twelve fragment hits were successfully determined at a high resolution. All twelve fragments were deeply inserted in the pocket and formed H-bonds with Asp73 and Thr165, and six fragments formed an additional H-bond with the backbone oxygen of Val71. Fragment screening further highlighted the capability of Asp73, Thr165 and Val71 to bind chemicals and provided diverse building blocks for the design of GyrB inhibitors.
Organizational Affiliation:
Research Center for Drug Discovery and Guangdong Provincial Key Laboratory of Chiral Molecule and Drug Discovery, School of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou 510006, China.