Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Imai, Y., Wakasugi, D., Suzuki, R., Kato, S., Sugisaki, M., Mima, M., Miyagawa, H., Endo, M., Fujimoto, N., Fukunaga, T., Kato, S., Kuroda, S., Takahashi, T., Kakinuma, H.(2022) Bioorg Med Chem Lett 79: 129050-129050
- PubMed: 36368497
- DOI: https://doi.org/10.1016/j.bmcl.2022.129050
- Primary Citation of Related Structures:
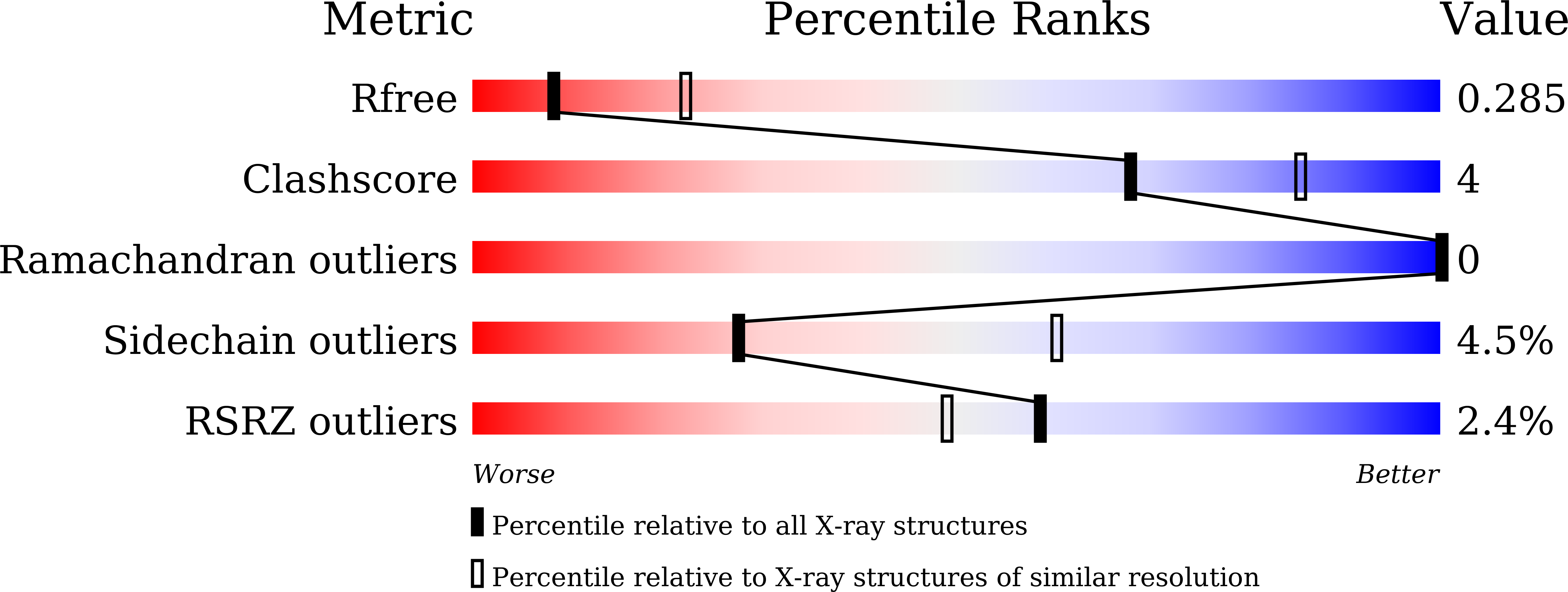
7YI7, 7YJC - PubMed Abstract:
Heparanase-1 (HPSE1) is an endo-β-d-glucuronidase that cleaves heparan sulfate proteoglycans into short-chain heparan sulfates (HS). The inhibition of HPSE1 has therapeutic potential for proteinuric diseases such as nephrotic syndrome because increased HPSE1 expression is associated with the loss of HS in the glomerular basement membrane, leading to the development of proteinuria. The present study examined the generation of a lead compound focusing on chemical structures with a sugar moiety, such as glycosides and sugar analogs, taking their physical properties into consideration. Compound 10, an exo-β-d-glucuronidase (GUSβ) inhibitor, was found to have a weak inhibitory activity against endo-β-d-glucuronidase HPSE1. A structure-activity relationship study using the X-ray co-crystal structure of 10 and HPSE1 resulted in 12a, which showed a more than 14-fold increase in HPSE1 inhibitory activity compared with that of 10. Compound 12a could be a novel lead compound for the development of a potent HPSE1 inhibitor.
Organizational Affiliation:
Medicinal Chemistry Laboratories, Taisho Pharmaceutical Co., Ltd., 1-403 Yoshino-cho, Kita-ku, Saitama 331-9530, Japan. Electronic address: y-imai@taisho.co.jp.