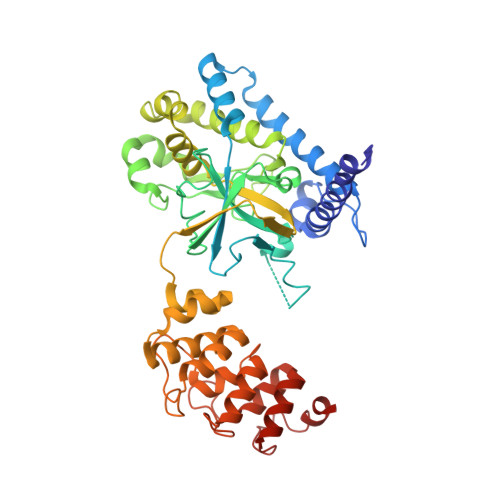
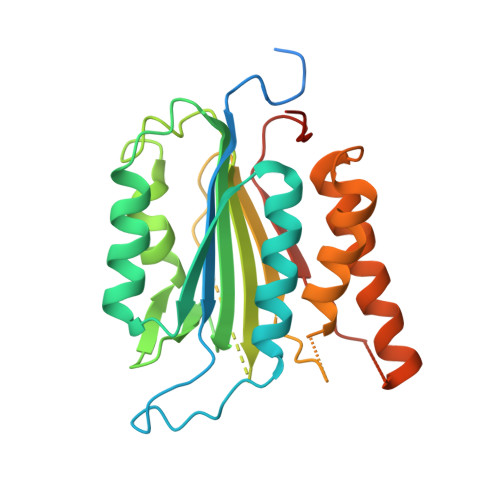
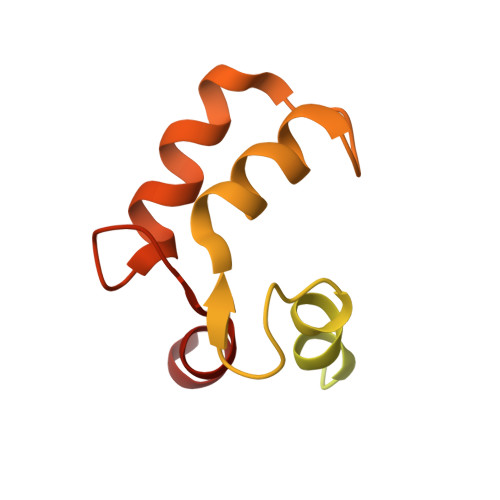
Calmodulin Binding Activates Chromobacterium CopC Effector to ADP-Riboxanate Host Apoptotic Caspases.
Liu, Y., Zeng, H., Hou, Y., Li, Z., Li, L., Song, X., Ding, J., Shao, F., Xu, Y.(2022) mBio 13: e0069022-e0069022
- PubMed: 35446120
- DOI: https://doi.org/10.1128/mbio.00690-22
- Primary Citation of Related Structures:
7WZS - PubMed Abstract:
Blocking host cell death is an important virulence strategy employed by many bacterial pathogens. We recently reported that Shigella flexneri inhibits host pyroptosis by delivering a type III secretion system (T3SS) effector OspC3 that catalyzes a novel arginine ADP-riboxanation modification on caspase-4/11. Here, we investigated the OspC3 homologue CopC from Chromobacterium violaceum, an opportunistic but sometimes deadly bacterial pathogen. CopC bears the same arginine ADP-riboxanase activity as OspC3, but with a different substrate specificity. Through proteomic analysis, we first identified host calmodulin (CaM) as a binding partner of CopC. The analyses additionally revealed that CopC preferably modifies apoptotic caspases including caspase-7, -8 and -9. This results in suppression of both extrinsic and intrinsic apoptosis programs in C. violaceum-infected cells. Biochemical reconstitution showed that CopC requires binding to CaM, specifically in the calcium-free state, to achieve efficient ADP-riboxanation of the caspases. We determined crystal structure of the CaM-CopC-CASP7 ternary complex, which illustrates the caspase recognition mechanism and a unique CaM-binding mode in CopC. Structure-directed mutagenesis validated the functional significance of CaM binding for stimulating CopC modification of its caspase substrates. CopC adopts an ADP-ribosyltransferase-like fold with a unique His-Phe-Glu catalytic triad, featuring two acidic residues critical for site-specific arginine ADP-riboxanation. Our study expands and deepens our understanding of the OspC family of ADP-riboxanase effectors. IMPORTANCE Programmed cell death is a suicidal defense mechanism for eukaryotes to combat pathogen infection. In the evolutionary arms race with the host, bacteria are endowed with ingenious tactics to block host cell death to facilitate their replication. Here, we report that the C. violaceum effector CopC ADP-riboxanates caspase-7/8/9, enabled by interacting with the host factor calmodulin, to block host cell apoptosis, illustrating a unique and sophisticated strategy adopted by the pathogen to counteract host defense.
Organizational Affiliation:
Graduate Program in Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.