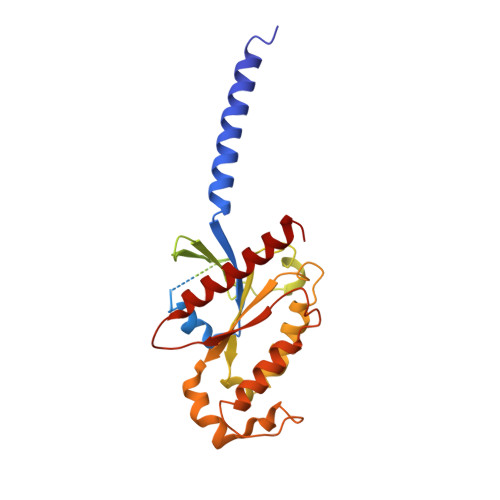
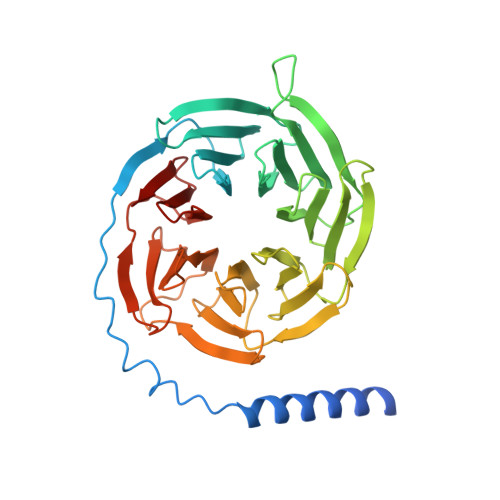
Cryo-EM structure of the human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand-binding specificity.
Heo, Y., Yoon, E., Jeon, Y.E., Yun, J.H., Ishimoto, N., Woo, H., Park, S.Y., Song, J.J., Lee, W.(2022) Elife 11
- PubMed: 35446253
- DOI: https://doi.org/10.7554/eLife.76823
- Primary Citation of Related Structures:
7WJ5 - PubMed Abstract:
Somatostatin is a peptide hormone that regulates endocrine systems by binding to G-protein-coupled somatostatin receptors. Somatostatin receptor 2 (SSTR2) is a human somatostatin receptor and is highly implicated in hormone disorders, cancers, and neurological diseases. Here, we report the high-resolution cryo-EM structure of full-length human SSTR2 bound to the agonist somatostatin (SST-14) in complex with inhibitory G (G i ) proteins. Our structural and mutagenesis analyses show that seven transmembrane helices form a deep pocket for ligand binding and that SSTR2 recognizes the highly conserved Trp-Lys motif of SST-14 at the bottom of the pocket. Furthermore, our sequence analysis combined with AlphaFold modeled structures of other SSTR isoforms provide a structural basis for the mechanism by which SSTR family proteins specifically interact with their cognate ligands. This work provides the first glimpse into the molecular recognition mechanism of somatostatin receptors and a crucial resource to develop therapeutics targeting somatostatin receptors.
Organizational Affiliation:
Department of Biochemistry, College of Life Science and Biotechnology, Yonsei University, Seoul, Republic of Korea.