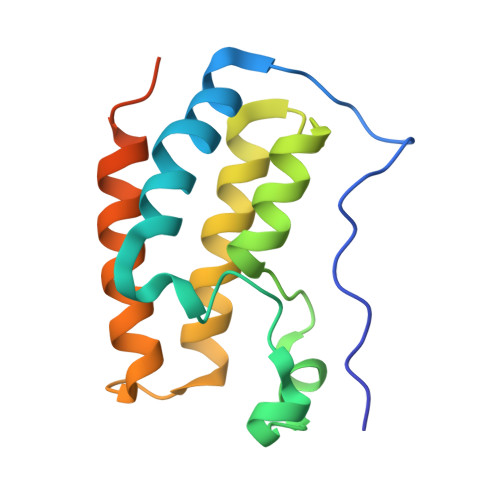
Binding of the SARS-CoV-2 envelope E protein to human BRD4 is essential for infection.
Vann, K.R., Acharya, A., Jang, S.M., Lachance, C., Zandian, M., Holt, T.A., Smith, A.L., Pandey, K., Durden, D.L., El-Gamal, D., Cote, J., Byrareddy, S.N., Kutateladze, T.G.(2022) Structure 30: 1224
- PubMed: 35716662
- DOI: https://doi.org/10.1016/j.str.2022.05.020
- Primary Citation of Related Structures:
7TUQ, 7TV0 - PubMed Abstract:
Emerging new variants of SARS-CoV-2 and inevitable acquired drug resistance call for the continued search of new pharmacological targets to fight the potentially fatal infection. Here, we describe the mechanisms by which the E protein of SARS-CoV-2 hijacks the human transcriptional regulator BRD4. We found that SARS-CoV-2 E is acetylated in vivo and co-immunoprecipitates with BRD4 in human cells. Bromodomains (BDs) of BRD4 bind to the C-terminus of the E protein, acetylated by human acetyltransferase p300, whereas the ET domain of BRD4 recognizes the unmodified motif of the E protein. Inhibitors of BRD4 BDs, JQ1 or OTX015, decrease SARS-CoV-2 infectivity in lung bronchial epithelial cells, indicating that the acetyllysine binding function of BDs is necessary for the virus fitness and that BRD4 represents a potential anti-COVID-19 target. Our findings provide insight into molecular mechanisms that contribute to SARS-CoV-2 pathogenesis and shed light on a new strategy to block SARS-CoV-2 infection.
Organizational Affiliation:
Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045, USA.