The ribosomal RNA processing 1B:protein phosphatase 1 holoenzyme reveals non-canonical PP1 interaction motifs.
Srivastava, G., Bajaj, R., Kumar, G.S., Gaudreau-Lapierre, A., Nicolas, H., Chamousset, D., Kreitler, D., Peti, W., Trinkle-Mulcahy, L., Page, R.(2022) Cell Rep 41: 111726-111726
- PubMed: 36450254
- DOI: https://doi.org/10.1016/j.celrep.2022.111726
- Primary Citation of Related Structures:
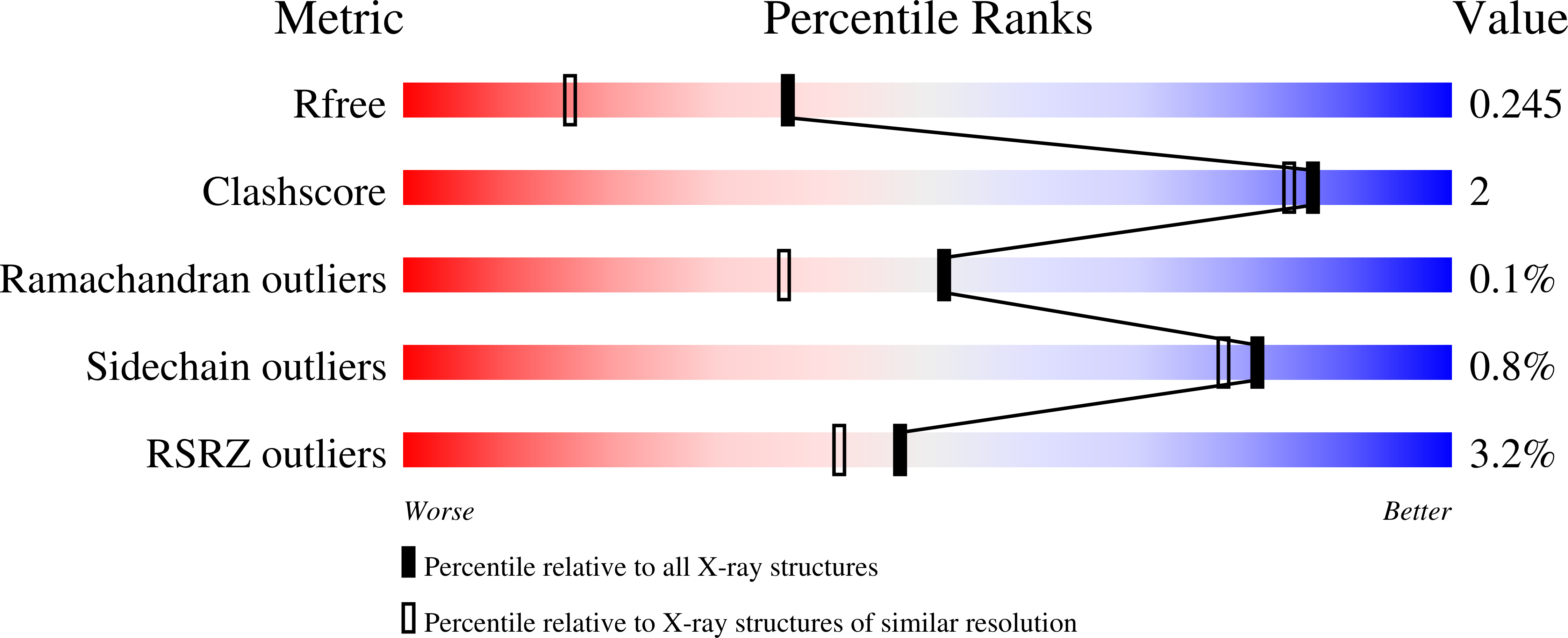
7T0Y - PubMed Abstract:
The serine/threonine protein phosphatase 1 (PP1) dephosphorylates hundreds of substrates by associating with >200 regulatory proteins to form specific holoenzymes. The major PP1 targeting protein in the nucleolus is RRP1B (ribosomal RNA processing 1B). In addition to selectively recruiting PP1β/PP1γ to the nucleolus, RRP1B also has a key role in ribosome biogenesis, among other functions. How RRP1B binds PP1 and regulates nucleolar phosphorylation signaling is not yet known. Here, we show that RRP1B recruits PP1 via established (RVxF/SILK/ΦΦ) and non-canonical motifs. These atypical interaction sites, the PP1β/γ specificity, and N-terminal AF-binding pockets rely on hydrophobic interactions that contribute to binding and, via phosphorylation, regulate complex formation. This work advances our understanding of PP1 isoform selectivity, reveals key roles of N-terminal PP1 residues in regulator binding, and suggests that additional PP1 interaction sites have yet to be identified, all of which are necessary for a systems biology understanding of PP1 function.
Organizational Affiliation:
Department of Molecular Biology and Biophysics, UConn Health, Farmington, CT 06030, USA.