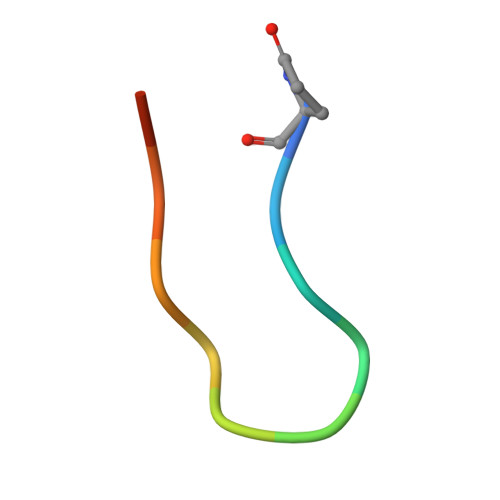
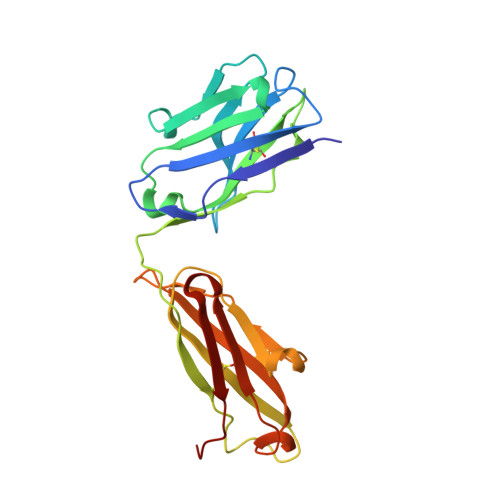
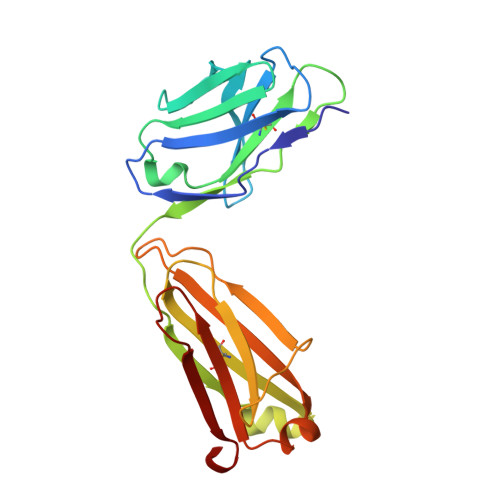
Discovery of a novel pseudo beta-hairpin structure of N-truncated amyloid-beta for use as a vaccine against Alzheimer's disease.
Bakrania, P., Hall, G., Bouter, Y., Bouter, C., Beindorff, N., Cowan, R., Davies, S., Price, J., Mpamhanga, C., Love, E., Matthews, D., Carr, M.D., Bayer, T.A.(2022) Mol Psychiatry 27: 840-848
- PubMed: 34776512
- DOI: https://doi.org/10.1038/s41380-021-01385-7
- Primary Citation of Related Structures:
7OW1, 7OXN - PubMed Abstract:
One of the hallmarks of Alzheimer's disease (AD) are deposits of amyloid-beta (Aβ) protein in amyloid plaques in the brain. The Aβ peptide exists in several forms, including full-length Aβ1-42 and Aβ1-40 - and the N-truncated species, pyroglutamate Aβ3-42 and Aβ4-42, which appear to play a major role in neurodegeneration. We previously identified a murine antibody (TAP01), which binds specifically to soluble, non-plaque N-truncated Aβ species. By solving crystal structures for TAP01 family antibodies bound to pyroglutamate Aβ3-14, we identified a novel pseudo β-hairpin structure in the N-terminal region of Aβ and show that this underpins its unique binding properties. We engineered a stabilised cyclic form of Aβ1-14 (N-Truncated Amyloid Peptide AntibodieS; the 'TAPAS' vaccine) and showed that this adopts the same 3-dimensional conformation as the native sequence when bound to TAP01. Active immunisation of two mouse models of AD with the TAPAS vaccine led to a striking reduction in amyloid-plaque formation, a rescue of brain glucose metabolism, a stabilisation in neuron loss, and a rescue of memory deficiencies. Treating both models with the humanised version of the TAP01 antibody had similar positive effects. Here we report the discovery of a unique conformational epitope in the N-terminal region of Aβ, which offers new routes for active and passive immunisation against AD.
Organizational Affiliation:
LifeArc, Centre for Therapeutics Discovery, Open Innovation Campus, Stevenage, UK. Preeti.Bakrania@lifearc.org.