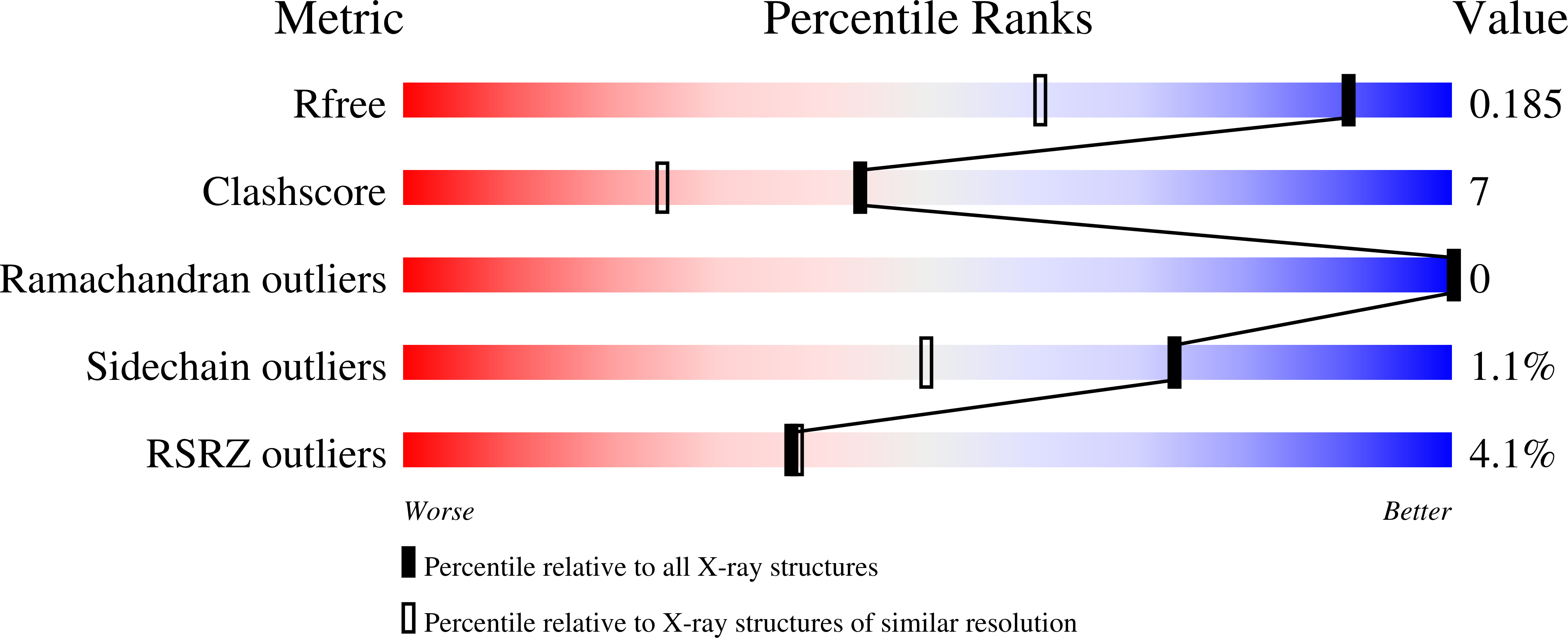
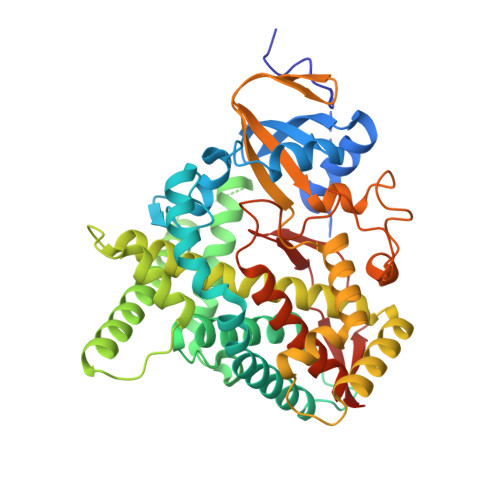
Design and synthesis of aryl-piperidine derivatives as potent and selective PET tracers for cholesterol 24-hydroxylase (CH24H)
Ikeda, S., Kajita, Y., Miyamoto, M., Matsumiya, K., Ishii, T., Nishi, T., Gay, S.C., Lane, W., Constantinescu, C.C., Alagille, D., Papin, C., Tamagnan, G., Kuroita, T., Koike, T.(2022) Eur J Med Chem 240: 114612