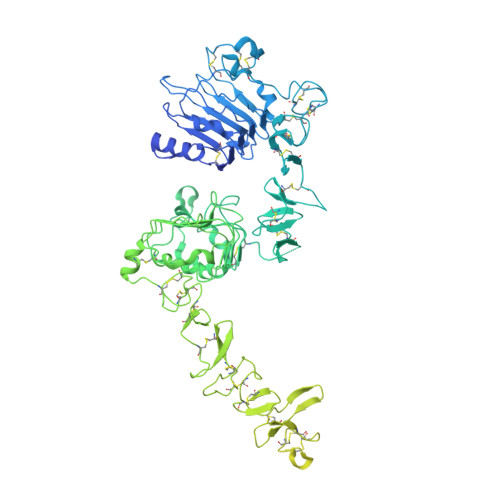
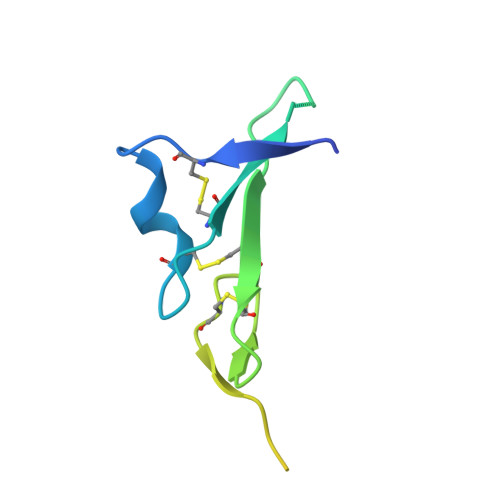
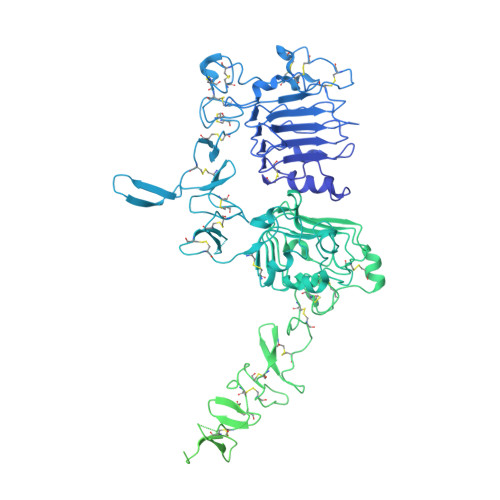
Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Diwanji, D., Trenker, R., Thaker, T.M., Wang, F., Agard, D.A., Verba, K.A., Jura, N.(2021) Nature 600: 339-343
- PubMed: 34759323
- DOI: https://doi.org/10.1038/s41586-021-04084-z
- Primary Citation of Related Structures:
7MN5, 7MN6, 7MN8 - PubMed Abstract:
Human epidermal growth factor receptor 2 (HER2) and HER3 form a potent pro-oncogenic heterocomplex 1-3 upon binding of growth factor neuregulin-1β (NRG1β). The mechanism by which HER2 and HER3 interact remains unknown in the absence of any structures of the complex. Here we isolated the NRG1β-bound near full-length HER2-HER3 dimer and, using cryo-electron microscopy, reconstructed the extracellulardomain module, revealing unexpected dynamics at the HER2-HER3 dimerization interface. We show that the dimerization arm of NRG1β-bound HER3 is unresolved because the apo HER2 monomer does not undergo a ligand-induced conformational change needed to establish a HER3 dimerization arm-binding pocket. In a structure of the oncogenic extracellular domain mutant HER2(S310F), we observe a compensatory interaction with the HER3 dimerization arm that stabilizes the dimerization interface. Both HER2-HER3 and HER2(S310F)-HER3 retain the capacity to bind to the HER2-directed therapeutic antibody trastuzumab, but the mutant complex does not bind to pertuzumab. Our structure of the HER2(S310F)-HER3-NRG1β-trastuzumab Fab complex reveals that the receptor dimer undergoes a conformational change to accommodate trastuzumab. Thus, similar to oncogenic mutations, therapeutic agents exploit the intrinsic dynamics of the HER2-HER3 heterodimer. The unique features of a singly liganded HER2-HER3 heterodimer underscore the allosteric sensing of ligand occupancy by the dimerization interface and explain why extracellular domains of HER2 do not homo-associate via a canonical active dimer interface.
Organizational Affiliation:
Cardiovascular Research Institute, University of California San Francisco, San Francisco, CA, USA.