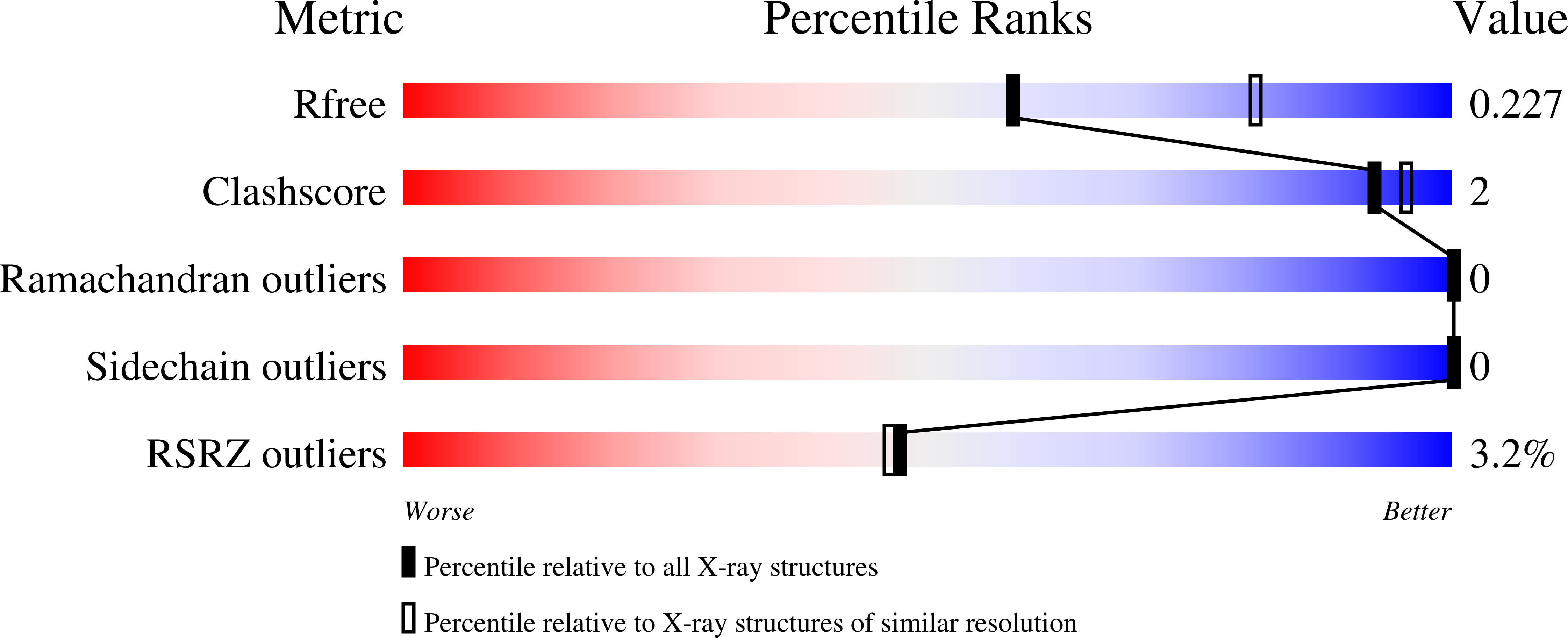
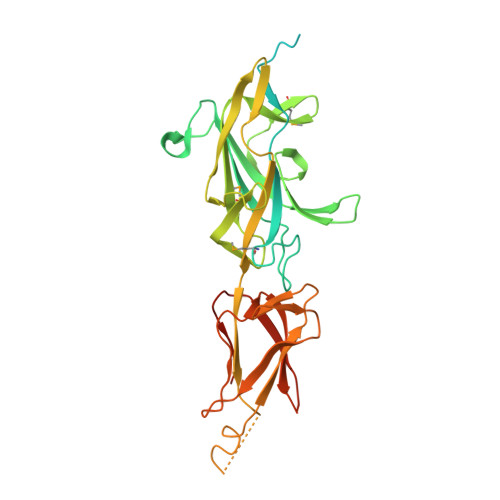
Crystal Structure of SrtB-anchored Collagen-binding Adhesin Fragment (residues 206-565) from Clostridioides difficile strain 630
Minasov, G., Shuvalova, L., Rosas-Lemus, M., Wiersum, G., Satchell, K.J.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.