Organism-specific differences in the binding of ketoprofen to serum albumin.
Czub, M.P., Stewart, A.J., Shabalin, I.G., Minor, W.(2022) IUCrJ 9: 551-561
- PubMed: 36071810
- DOI: https://doi.org/10.1107/S2052252522006820
- Primary Citation of Related Structures:
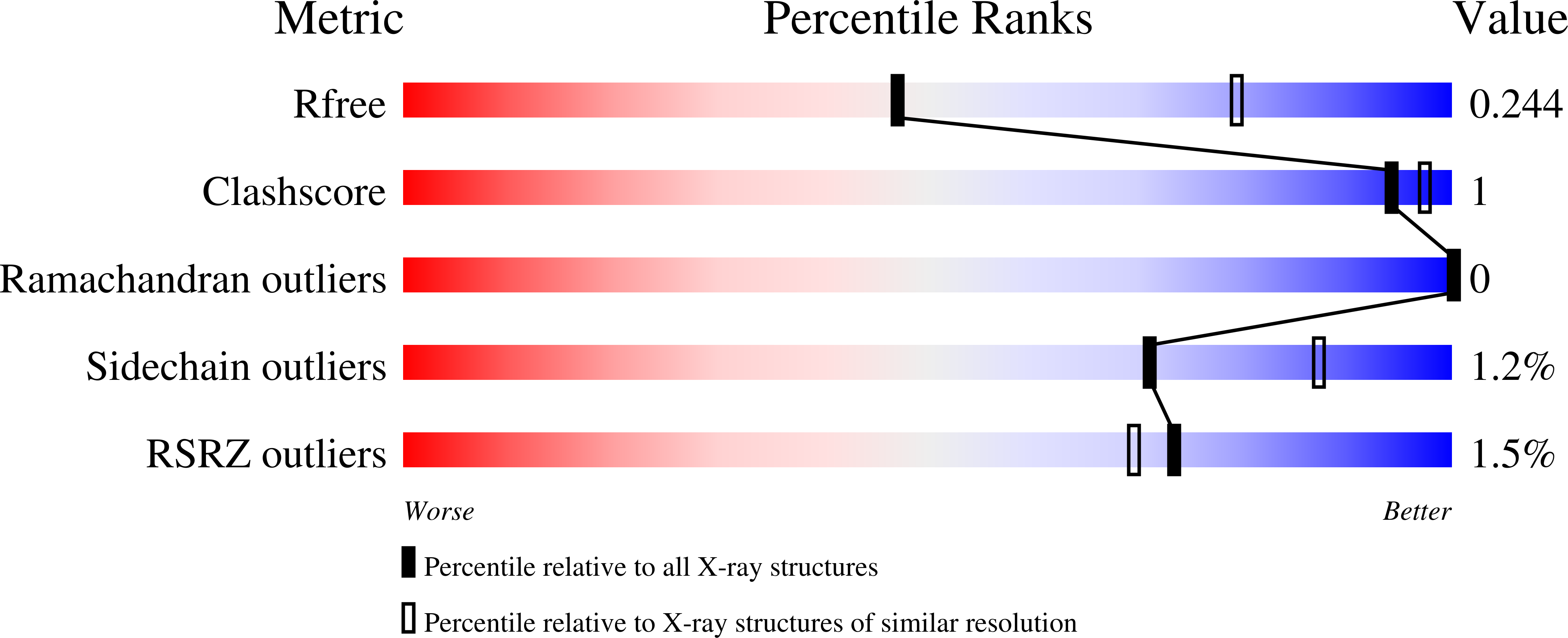
7JWN - PubMed Abstract:
Serum albumin is a circulatory transport protein that has a highly conserved sequence and structure across mammalian organisms. Its ligand-binding properties are of importance as albumin regulates the pharmacokinetics of many drugs. Due to the high degree of structural conservation between mammalian albumins, nonhuman albumins such as bovine serum albumin or animal models are often used to understand human albumin-drug interactions. Ketoprofen is a popular nonsteroidal anti-inflammatory drug that is transported by albumin. Here, it is revealed that ketoprofen exhibits different binding-site preferences when interacting with human serum albumin compared with other mammalian albumins, despite the conservation of binding sites across species. The reasons for the observed differences were explored, including identifying ketoprofen binding determinants at specific sites and the influence of fatty acids and other ligands on drug binding. The presented results reveal that the drug-binding properties of albumins cannot easily be predicted based only on a complex of albumin from another organism and the conservation of drug sites between species. This work shows that understanding organism-dependent differences is essential for assessing the suitability of particular albumins for structural or biochemical studies.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, 1340 Jefferson Park Avenue, Charlottesville, VA 22908, USA.