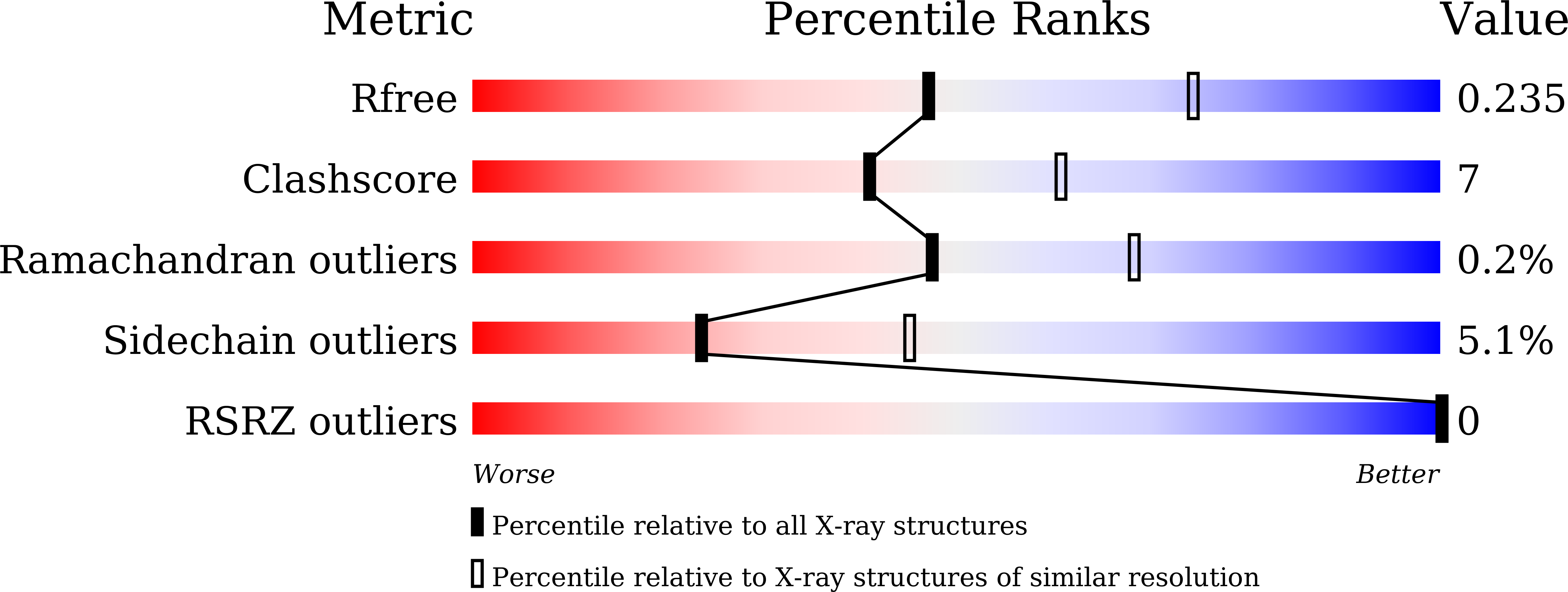
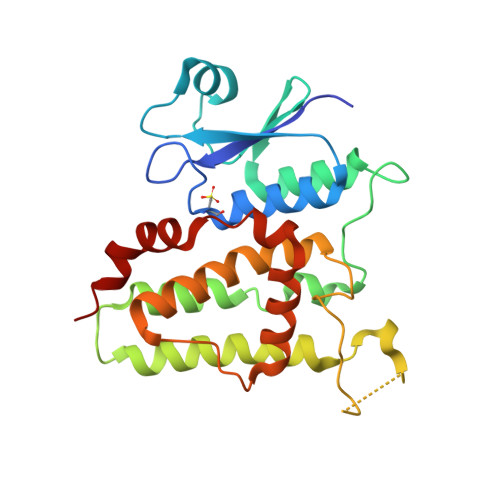
Comparative kinetic analysis of ascorbate (Vitamin-C) recycling dehydroascorbate reductases from plants and humans.
Das, B.K., Kumar, A., Sreekumar, S.N., Ponraj, K., Gadave, K., Kumar, S., Murali Achary, V.M., Ray, P., Reddy, M.K., Arockiasamy, A.(2021) Biochem Biophys Res Commun 591: 110-117
- PubMed: 35007834
- DOI: https://doi.org/10.1016/j.bbrc.2021.12.103
- Primary Citation of Related Structures:
7F8R, 7F8S - PubMed Abstract:
Ascorbate is an important cellular antioxidant that gets readily oxidized to dehydroascorbate (DHA). Recycling of DHA is therefore paramount in the maintenance of cellular homeostasis and preventing oxidative stress. Dehydroascorbate reductases (DHARs), in conjunction with glutathione (GSH), carry out this vital process in eukaryotes, among which plant DHARs have garnered considerable attention. A detailed kinetic analysis of plant DHARs relative to their human counterparts is, however, lacking. Chloride intracellular channels (HsCLICs) are close homologs of plant DHARs, recently demonstrated to share their enzymatic activity. This study reports the highest turnover rate for a plant DHAR from stress adapted Pennisetum glaucum (PgDHAR). In comparison, HsCLICs 1, 3, and 4 reduced DHA at a significantly lower rate. We further show that the catalytic cysteine from both homologs was susceptible to varying degrees of oxidation, validated by crystal structures and mass-spectrometry. Our findings may have broader implications on crop improvement using pearl millet DHAR vis-à-vis discovery of cancer therapeutics targeting Vitamin-C recycling capability of human CLICs.
Organizational Affiliation:
Membrane Protein Biology Group, International Centre for Genetic Engineering and Biotechnology, Aruna Asaf Ali Marg, New Delhi, 110067, India; Southern California Institute for Research and Education, Veterans Affairs Long Beach Health Care System, 5901 E. 7th Street Long Beach, CA, 90822, USA.