Cysteine Engineering of an Endo-polygalacturonase from Talaromyces leycettanus JCM 12802 to Improve Its Thermostability.
Wang, S., Meng, K., Su, X., Hakulinen, N., Wang, Y., Zhang, J., Luo, H., Yao, B., Huang, H., Tu, T.(2021) J Agric Food Chem 69: 6351-6359
- PubMed: 34043362
- DOI: https://doi.org/10.1021/acs.jafc.1c01618
- Primary Citation of Related Structures:
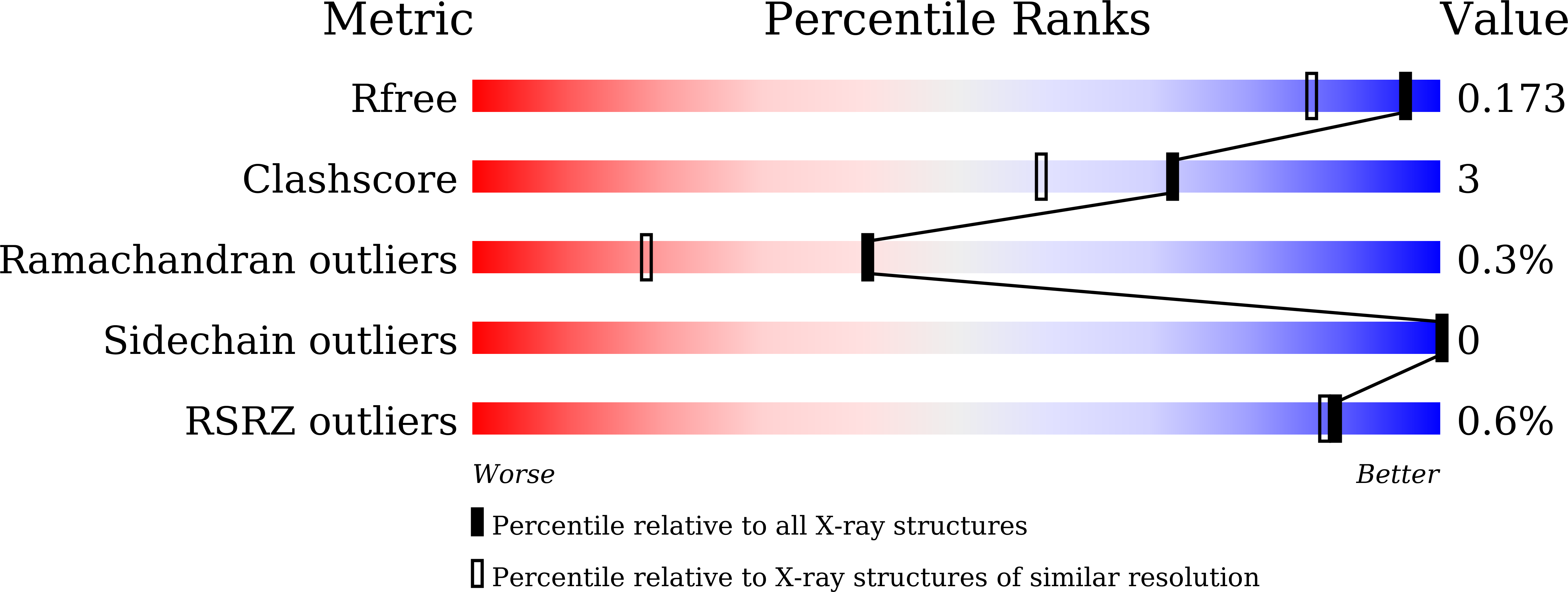
7E56 - PubMed Abstract:
Thermostable enzymes have many advantages for industrial applications. Therefore, in this study, computer-aided design technology was used to improve the thermostability of a highly active endo-polygalacturonase from Talaromyces leycettanus JCM12802 at an optimal temperature of 70 °C. The melting temperature and specific activity of the obtained mutant T316C/G344C were increased by 10 °C and 36.5%, respectively, compared with the wild-type enzyme. The crystal structure of the T316C/G344C mutant showed no formation of a disulfide bond between the introduced cysteines, indicating a different mechanism than the conventional mechanism underlying improved enzyme thermostability. The cysteine substitutions directly formed a new alkyl hydrophobic interaction and caused conformational changes in the side chains of the adjacent residues Asn315 and Thr343, which in turn caused a local reconstruction of hydrogen bonds. This method greatly improved the thermostability of the enzyme without affecting its activity; thus, our findings are of great significance for both theoretical research and practical applications.
Organizational Affiliation:
State Key Laboratory of Animal Nutrition, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing 100193, China.