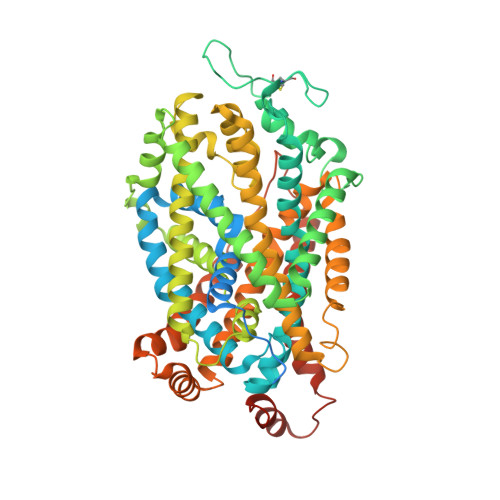
Chemical and structural investigation of the paroxetine-human serotonin transporter complex.
Coleman, J.A., Navratna, V., Antermite, D., Yang, D., Bull, J.A., Gouaux, E.(2020) Elife 9
- PubMed: 32618269
- DOI: https://doi.org/10.7554/eLife.56427
- Primary Citation of Related Structures:
6VRH, 6VRK, 6VRL, 6W2B, 6W2C - PubMed Abstract:
Antidepressants target the serotonin transporter (SERT) by inhibiting serotonin reuptake. Structural and biochemical studies aiming to understand binding of small-molecules to conformationally dynamic transporters like SERT often require thermostabilizing mutations and antibodies to stabilize a specific conformation, leading to questions about relationships of these structures to the bonafide conformation and inhibitor binding poses of wild-type transporter. To address these concerns, we determined the structures of ∆N72/∆C13 and ts2-inactive SERT bound to paroxetine analogues using single-particle cryo-EM and x-ray crystallography, respectively. We synthesized enantiopure analogues of paroxetine containing either bromine or iodine instead of fluorine. We exploited the anomalous scattering of bromine and iodine to define the pose of these inhibitors and investigated inhibitor binding to Asn177 mutants of ts2-active SERT. These studies provide mutually consistent insights into how paroxetine and its analogues bind to the central substrate-binding site of SERT, stabilize the outward-open conformation, and inhibit serotonin transport.
Organizational Affiliation:
Vollum Institute, Oregon Health & Science University, Portland, United States.